INTRODUCTION
High-throughput genomic technologies have provided opportunities for increased understanding of nutritional modulation of gene and proteins expression 1) (2. However, this approach has limitations, such as evaluating slight changes and limited access to specific tissues, especially in patient samples composed of healthy volunteers.
According to the literature, some genes are expressed in tissue-specific manner 2),(3, consequently, it is necessary to access to the biological material. In cases of patients who will undergo surgery or biopsy, collecting tissue is considered as feasible. Otherwise, without this procedure, collection of internal organ samples such as visceral and adipose tissue is less probable, especially for ethical reasons 4) (5.
A minimally invasive method for the study of health-illness transitions is the collection of blood obtained by using the standard venipuncture technique. Peripheral blood mononuclear cells (PBMC) are exposed to all physiological changes, including the change of nutrient concentrations according to their ingestion and absorption 1) (6) (7) (8.
The evaluation of PBMC for transcriptome analysis has been successfully used to discriminate potential diseases to healthy populations 7) (8) (9) (10. Despite PBMC is easily obtainable compared to adipose tissue, liver and muscle, literature has shown conflicting results, and assessing a single tissue could not be enough to fully understand changes in the body 1) (2) (3. In this study, we aimed to determine whether targets gene expression in the PBMC is able to reflect the same expression profile in the subcutaneous white adipose tissue in patients with severe obesity.
MATERIALS AND METHODS
This transversal study enrolled patients from a mixed population 11, 35 obese grade III individuals (OG) with a BMI > 40 kg/m2 and ten eutrophic individuals (BMI ranging from 18.5 kg/m² to 24.9 kg/m²) as a CG. The sample size was defined by convenience, according to the number of patients treated at the hospital where they were selected. The control individuals underwent surgery of an umbilical hernia (incisional or epigastric) or gallstones without acute cholecystitis, both from the Clinical Hospital of Ribeirão Preto Medical School, of the University of São Paulo. After a thorough introduction to the study, all participants gave their written informed consent to participate. This study was approved by the Ethical Committee of the institution (Certificate of Presentation for Ethical Consideration - CAAE: 18973913.0.0000.5440).
Anthropometric and body composition data, blood and adipose tissue were collected from both groups. Weight (kg), height (m) and BMI (kg/m²) were determined at recruitment using standardized protocols. Body composition was assessed by bioelectrical impedance through Quantum BIA 101 q-RJL Systems analyzer (Clinton Township, MI, USA). Adipose tissue was collected during the surgery procedure or by biopsy in the right upper quadrant region above umbilicus cicatrix.
GENE EXPRESSION
RNA was extracted from PBMC and white adipose tissue for gene expression analysis of gene involved in adipogenesis and energy metabolism (PLIN1, ADRB3 and PPARG2, UCP1, UCP2 and UCP3).
RNA was extracted from samples of subcutaneous white adipose tissue and PBMC using the phenol-chloroform extraction method modified by Chomczynski & Sacchi (1987) 12. The real-time quantitative polymerase chain reaction (RT-qPCR) method was prepared as described above. RNA was reverse transcribed using a high capacity cDNA reverse transcription with a RNase inhibitor kit (Applied Biosystems-Life Technologies, Carlsbad, CA). Quantitative PCR was performed in triplicate on the ABI 7500 Fast Plate (Applied Biosystems). The RT-qPCR reactions were assembled based on the TaqMan Universal PCR Master Mix protocol (Applied Biosystems), using the following genes: UCP1 (Hs00222453_m1), UCP2 (Hs01075227_m1), UCP3 (Hs01106052_m1), ADRB3 (Hs00609046_m1), PLIN1 (Hs00160173_m1) and PPARG2 (Hs01115513_m1). GAPDH (Hs02758991_m1) and ACTB (Hs99999903_m1) were used as the calibrator genes. Quantitative real-time PCR was performed essentially as described and followed the MIQE guidelines 13. The fold change in gene expression in PBMCs and white adipose tissue is quantified relative to the use of the control group as calibrator in this study.
STATISTICAL ANALYSIS
Descriptive statistics consisted of mean values and standard deviation. The normality of data was verified by the Shapiro-Wilk test. The Mann-Whitney test was used to compare gene expression between tissues. Statistical significance was set at p < 0.05, and all analyses were performed in the Statistical Package software for Social Sciences (SPSS version 20.0 Inc. Chicago. IL).
RESULTS
Anthropometric and body composition data are described in table I. We found, as expected, that weight, BMI, waist circumference (WC), lean mass (LM, kg), fat mass (FM, kg) and fat proportion (%) were increased in patients with obesity grade III compared with eutrophic (p < 0.05).
Table I Anthropometric and body composition data of obese patients and normal weight individuals
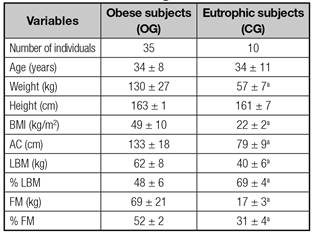
Unpaired t test. BMI: Body mass index; AC: Abdominal circumference; LBM: Lean body mass; FM: Fat masss. ap < 0.05 compared to OG.
Figure 1 shows the relative gene expression in blood and adipose tissue of target genes. The present study indicates that the UCP1 gene was slightly expressed in evaluated tissues, and PLIN1, ADRB3, PPARG2 and UCP3 were upregulated only in samples of subcutaneous adipose tissue in patients with severe obesity. On the other hand, UCP2 expression was upregulated in blood cells in the same patients (Fig. 1A). Statistical analysis showed some outliers, therefore, a new analysis was performed by removing possible confounding factors, but results remained the same. In order to avoid reducing the number of samples, original values, which are featured by asterisk in the graphs, are maintained (Figs. 1A-F).
DISCUSSION
In the current study, we demonstrated that the expression of genes involved in energy and lipid metabolism is different in white subcutaneous adipose tissue when compared to mononuclear cells from peripheral blood of patients with severe obesity. We observed that expression of UCP2 was upregulated in blood samples, whereas UCP3, PLIN1, PPARG2 and ADRB3 were upregulated in white adipose tissue.
The main purpose of this study was to identify similarities between gene expression in PBMCs and white adipose tissue, and validate the use of peripheral blood considering easy access, availability and handling. However, all genes evaluated behaved differently among tissues, suggesting that it is not possible to obtain a reflection of all changes in specific tissues from this biological material.
Dietary interventions for weight loss have shown an impact of several genes on subcutaneous adipose tissue 14) (15) (16, although intra- and inter-individual variation has not been fully studied, especially in blood samples. Brattbakk et al. (2013) 3 showed that the correlation between gene expression in peripheral blood and adipose tissue varies according to gene function, suggesting that more studies about this relation are needed for better understanding the impact of diet on the body. In the same way, a recent study showed that the expression of some genes in white adipose tissue and blood can be used for nutritional studies as predictive markers expansion of white adipose tissue 17. Likewise, a study by Diaz-Rua et al. (2015) 18 argues that the analysis of gene expression in PBMCs allows the detection of physiological deviations induced by diet.
Nutrigenomic studies in humans involving the regulation of gene transcription in different tissues are still scarce, first, because this is a relatively new scientific field, and also, because it is not being fully integrated into clinical practice. Moreover, costs for the implementation of this technology are still significant, especially in numerous approaches involving populations 1.
Additionally, most studies in this area aim to identify whether changes in gene transcription can be detected after changes in food intake, suggesting that food and nutrients can cause a significant impact on the ability of adaptive cellular response to changes in the gene expression pattern 1) (19. Nevertheless, for this analysis, specific tissue fragments, which are often difficult to access, are needed to evaluate the best response induced by intervention.
The present study found that some genes are more significantly expressed in white adipose tissue; however, the UCP2 gene, an important regulator of energy metabolism, is more expressed in blood cells. Alternatively, Mello et al. (2012) 2 emphasize that gene expression of PBMC after a dietary intervention can be used to examine the gene response related to the PPAR path and cholesterol metabolism, suggesting that PBMCs appear to be promising for their use in nutritional genomics.
Other studies suggest that gene expression in peripheral blood may be more variable when compared to other tissues 3) (20. However, literature has also shown that gene expression of the same individual at the right time is stable in blood cells 7) (8) (21) (22. These factors suggest that peripheral blood could be used in transcriptome analysis studies to verify responses to dietary interventions derived from the same individual over time 3.
The main limitations of this study relate to the sample size and the reduced number of genes evaluated. However, our research group has achieved positive results for the analysis of gene expression in small numbers of patients 16) (23, although it would be important to assess the whole transcriptome comparing PBMCs and specific tissue to detect biomarkers for disorders or physiological changes related to the pathogenesis of diseases. Thus, having a gene signature from a biological material easily accessible, such as blood cells, would be important so that this approach could identify specific nutritional problems and chances for disease.
In conclusion, the current study demonstrated that the expression of genes involved in energy expenditure (UCP3) and lipid metabolism (PLIN1, PPARG2 and ADRB3) are downregulated in peripheral blood of patients with severe obesity. Moreover, UCP2 is upregulated in PBMCs, suggesting that the expression of this gene in peripheral blood could be used as a marker present in the material easily accessed and handled.












 Curriculum ScienTI
Curriculum ScienTI


