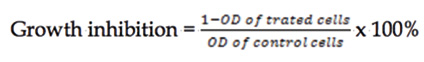INTRODUCTION
The heat shock proteins (Hsp) are induced in response to heat shock and other conditions such as chemical or physical stress conditions; these are encoded by the Hsp 27, 28, 40, 70, 90 and 110 genes. These proteins possess the property of modifying other proteins, their functions and these interact to alter their normal functioning. They are highly efficient in synthesis of proteins in response to stress and have highly efficient stabilisation effect thus maintaining the normal homeostasis by transcriptional activation, mRNA stabilisation and translation.1) Several studies have proved the role of Hsp70 and 90 in various biochemical and physiological process such as regulation of apoptosis via the apoptotic pathway, regulation for release and transport of mitochondrial cytochrome c, regulating the tumour necrosis factor (TNF) and its related apoptoticmoieties to respective receptors.2) Hsp 70 and 90 are highly expressed in cancer that are drug resistant and are often observed in patients suffering from drug resistance to chemotherapeutic agents.3 Thus, these proteins have become logical and popular targets fordevelopment of anticancer agents. On this front very few drugs are available with Geldanamycin, Macbecin and Radicicolas only compounds available for inhibiting the Hsp90. Whereas, Apoptozole, dibenzyladenosine analogues, Pifithrin, several pyrimidine analogues and flavonoids are found to inhibit Hsp70, but all these compounds are under investigation or in clinical phase.4) It has been well observed that overexpression of these heat shock proteins is one of the factors in progression of prostate cancer. Thus development of compounds that can inhibit these proteins could possibly lead to control over prostatic adenocarcinoma.5),(6) This situation inspired us for development of some newer derivatives as inhibitors of Hsp70 and Hsp90 for treatment of prostate cancer.
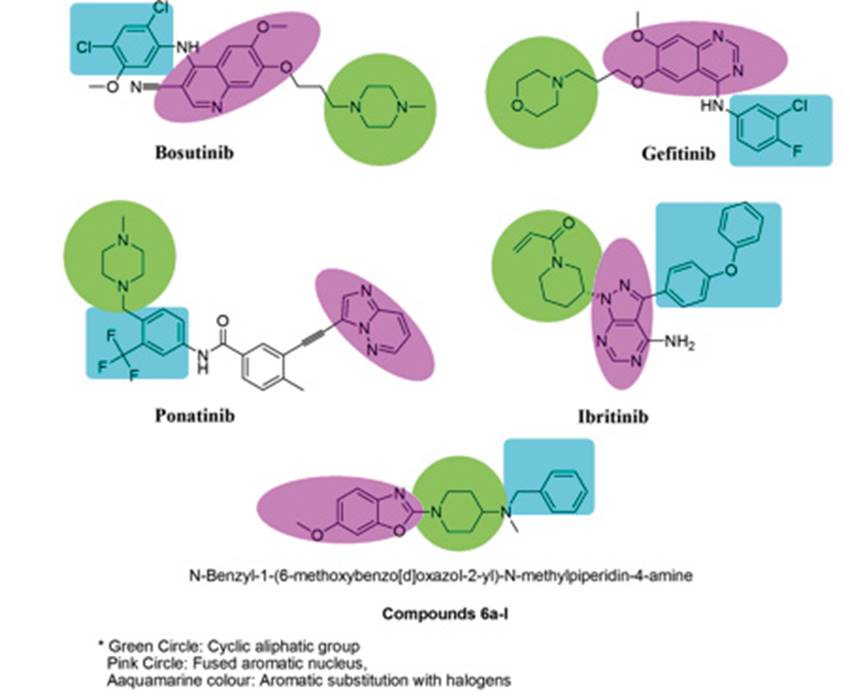
Figure 1: Anticancer drugs with hybrid nature and possess common three pharmacophore features as cyclic aliphatic group with substitution, fused aromatic nucleus and the aryl substitutions generally acting as linker.
Several anticancer compounds are reported to have inhibitory activity towards the Hsp90 and Hsp70. On analysing these compounds it was found that these molecules have several common structural features and belongs to the ‘nib’ class of drugs. Compounds such as Bosutinib, Gefitinib, Ponatinib, Imatinib, Ceitinib, Afitinib and Ibrutinib have inhibitory action on the Hsp90 and 70.7) (8) (9) (10 It was observed that these compounds have several common structural features such as fused ring structures or have an aliphatic cyclic moiety with heteroatom and substituted aryl ring (Figure 1). Along with these structural features it was observed that these compounds are hybrid in nature, formed from two or more biologically active chemical moieties. Benzoxazole are reported to possess antitumor activity and can be synthesised by various routes, similarly piperidine is also reported to have anticancer activity.11) (12) (13) Based on these observations we designed novel hybrid molecules of benzoxazole and piperidine with structural features similar to the cancer therapeutic agents. Benzoxazole was linked with substituted piperidine to yieldtwelve derivatives with different substitutions on the aryl ring (Scheme 1).

Scheme 1 Reagents and conditions for the synthesis of 6a-l: a) Dimethyl formamide (DMF), Potassium carbonate (K2CO3), room temperature, 6 h; b) Sodium methoxide (CH3Na), Methanol, reflux, 12 h; c) Potassium Iodide (KI), Tetrahydrofuran (THF), Trifluroacetic acid, DCM, room temperature, 7 h; d) K2CO3, DMF, 4 h.
MATERIAL AND METHODS
Chemicals and solvents for this work were obtained from Alfa Aeser, UK and Sigma Aldrich, USA unless otherwise mentioned. Thiele’s tube melting-point apparatus was used to determine the melting points (mp) for all the compounds within open capillaries and was recorded without corrections. Infrared (IR) spectra were developed and created on JASCO FT/IR-4000 spectrophotometer (Jasco, USA) with KBr pellets used for compounds. Proton NMR (1H) spectra were recorded on a Bruker Advance-II 400 Spectrometer on 400 MHz using tetramethylsilane (TMS) as internal standard. Chemical shift values for the synthetic derivatives were recorded as ? (parts per million), coupling constant value Jwas measured in hertz (Hz). The peaks are presented as s = singlet, d = doublet, t = triplet, dd = double doublet, m = multiplet. In similar way 13C NMR were recorded. Thin Layer Chromatographic (TLC) method was used to determine the purity of compounds and progress of reaction using Merck, silica gel, HF254-361, type 60, 0.25 mm. Electron Spray Ionisation Mass Spectrometry (ESI-MS) were recorded on Waters HPLC system with Q-Time of Flight LC-MS spectrometry (Waters-Micromass LC-MS, US).
Chemistry
The title compounds N-substituted benzyl-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine6a-l were obtained starting from benzoxazole in four step synthetic procedure, these procedures are discussed below as follows;
Synthesis of tert-butyl 1-(6-chlorobenzo[d]oxazol-2-yl)piperidin-4-yl(methyl)carbamate3
A mixture of 2,6-dichlorobenzo[d]oxazole1 (1.0 mmol) and tert-butyl methyl(piperidin-4-yl)carbamate 2 (1.0 mmol) were stirred in DMF (5 mL) in presence of potassium carbonate for about 6 hours at room temperature, completion of reaction was monitored on TLC. The resulting compound tert-butyl 1-(6-chlorobenzo[d]oxazol-2-yl)piperidin-4-yl(methyl) carbamate3 was isolated as off white ppt in 77% yield. Mp: 230-232 ºC; 1H-NMR (CDCl3): ? 1.49 (s, 9H, Boc-CH3), 1.60 (m, 2H, Piperidine-H), 1.89 (m, 2H, Piperidine-H), 2.3 (m, 2H, Piperidine-H), 2.5 (m, 2H, Piperidine-H), 2.9 (s, 3H, N-CH3), 3.72 (m, 2H, methylene-H), 7.20 (m, 1H, Ar-H), 7.85 (m, 1H, Ar-H); Anal. Calcd for C18H24ClN3O3; C, 59.09; H, 6.61; Cl, 9.69; N, 11.49; O, 13.12; MS (ESI) m/z: 365.1506.
Synthesis of tert-butyl 1-(6-methoxybenzo[d]oxazol-2-yl)piperidin-4-yl(methyl)carbamate 4
Compound 3 (10 mmol) was taken in methanol (100 mL) and allowed to solubilise, to this was added sodium methoxide (15 mmol) and refluxed for 7 h. The mixture so obtained was cooled to room temperature and excess of solvent was removed under vacuo. The obtained residue was extracted with ethyl acetate and dried over sodium sulphate to obtain the white coloured amorphous product 4 with 62 % yield. Mp: 292-294 ºC; 1H-NMR (CDCl3): ? 1.45 (s, 9H, Boc-CH3), 1.62 (m, 2H, Piperidine-H), 1.88 (m, 2H, Piperidine-H), 2.2 (m, 2H, Piperidine-H), 2.4 (m, 2H, Piperidine-H), 2.7 (s, 3H, N-CH3), 3.12 (s, 3H, OCH3), 3.79 (m, 2H, methylene-H), 7.21 (m, 1H, Ar-H), 7.80 (m, 1H, Ar-H); Anal. Calcd for C19H27N3O4; C, 63.14; H, 7.53; N, 11.63; O, 17.71; MS (ESI) m/z: 361.2.
Synthesis of 1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 5
Compound 4 (10 mmol) was taken in tetrahydrofuran (100 mL) and mixed to form a uniform mixture, to this was added potassium iodide (10 mmol) and refluxed for 12 h. The mixture so obtained was cooled to room temperature and excess of solvent was removed under vacuo. The obtained residue was treated with trifluoroacetic acid in dichloroethane to obtain deprotected amine5 with 71 % yield. Mp: 167-169 ºC; 1H-NMR (CDCl3): ? 1.11 (m, 2H, Piperidine-H), 1.34 (m, 2H, Piperidine-H), 2.1 (m, 2H, Piperidine-H), 2.6 (m, 2H, Piperidine-H), 2.7 (s, 3H, N-CH3), 3.12 (s, 3H, OCH3), 3.79 (m, 2H, methylene-H), 7.25 (m, 1H, Ar-H), 7.88 (m, 1H, Ar-H), 8.4 (s, 1H, NH); Anal. Calcd for C14H19N3O2; C, 64.35; H, 7.33; N, 16.08; O, 12.25; MS (ESI) m/z: 261.15.
General procedure for the synthesis of various N-substituted benzyl-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6a-l
In the last step compound 5 (0.2 mmol) was added to DMF (10 mL), to this solution anhydrous potassium carbonate (0.30 mmol) followed by various substituted benzyl chloride (0.2 mmol). This solution was stirred for 4 h at room temperature, on completion of reaction on the TLC water was added. The compound was extracted with DCM and dried over sodium sulphate, this was dried in vacuo to provide with final derivatives 6a-l.
N-Benzyl-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6a
59 % yield. Mp: 157-159 ºC; 1H-NMR (CDCl3): ? 1.43 (m, 2H, Piperidine-H), 1.68 (m, 2H, Piperidine-H), 2.65 (s, 3H, N-CH3), 2.69 (m, 1H, Piperidine-H), 2.7 (m, 2H, Piperidine-H), 2.8 (m, 2H, Piperidine-H), 3.62 (s, 2H, methylene-H), 3.73 (s, 3H, OCH3), 7.06 (m, 2H, Ar-H), 7.1 (m, 1H, Ar-H), 7.2 (m, 2H, Ar-H), 7.25 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H); 13C NMR spectral data (25.15 MHz, CDCl3): ?136.5 (C-2), 143.4 (C-4), 149.9 (C-5), 95.5 (C-6), 158.3(C-7), 112.9(C-8), 119.7(C-9), 49.6(C-11), 31.8(C-12), 51.3(C-13), 31.8(C-14), 49.6(C-15), 55.5(C-17), 42.7(C-19), 57.5 (C-20), 136.7(C-21), 128.0 (C-22, C-26), 128.6(C-23,C-25), 128.9 (C-24) Anal. Calcd for C21H25N3O2; C, 71.77; H, 7.17; N, 11.96; O, 9.10; MS (ESI) m/z: 351.1947.
N-(4-chlorobenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine6b
51 % yield. Mp: 151-153 ºC; 1H-NMR (CDCl3): ? 1.44 (m, 2H, Piperidine-H), 1.67 (m, 2H, Piperidine-H), 2.67 (s, 3H, N-CH3), 2.68 (m, 1H, Piperidine-H), 2.75 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.08 (m, 2H, Ar-H), 7.29 (m, 2H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H); 13C NMR spectral data (25.15 MHz, CDCl3): ? 136.5 (C-2), 143.4 (C-4), 149.2 (C-5), 95.9 (C-6), 158.4(C-7), 112.4(C-8), 119.7(C-9), 49.3(C-11), 31.5(C-12), 51.3(C-13), 31.8(C-14), 49.7(C-15), 55.7(C-17), 42.4(C-19), 57.3 (C-20), 136.5(C-21), 129.6 (C-22, C-26), 131.8(C-23,C-25), 124.2 (C-24) Anal. Calcd for C21H24ClN3O2; C, 65.36; H, 6.27; Cl, 9.19; N, 10.89; O, 8.29; MS (ESI) m/z: 385.1557.
N-(4-bromobenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6c
55 % yield. Mp: 154-156 ºC; 1H-NMR (CDCl3): ? 1.39 (m, 2H, Piperidine-H), 1.59 (m, 2H, Piperidine-H), 2.59 (s, 3H, N-CH3), 2.68 (m, 1H, Piperidine-H), 2.75 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.11 (m, 2H, Ar-H), 7.30 (m, 2H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H); ); 13C NMR spectral data (25.15 MHz, CDCl3): ? 136.5 (C-2), 143.4 (C-4), 149.2 (C-5), 95.9 (C-6), 158.4(C-7), 112.4(C-8), 119.7(C-9), 49.3(C-11), 31.5(C-12), 51.3(C-13), 31.8(C-14), 49.7(C-15), 55.7(C-17), 42.4(C-19), 57.3 (C-20), 136.5(C-21), 129.6 (C-22, C-26), 131.8(C-23,C-25), 124.2 (C-24); Anal. Calcd for C21H24BrN3O2; C, 58.61; H, 5.62; Br, 18.57; N, 9.76; O, 7.44; MS (ESI) m/z: 429.1052
N-(4-fluorobenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6d
61 % yield. Mp: 150-152 ºC; 1H-NMR (CDCl3): ? 1.43 (m, 2H, Piperidine-H), 1.66 (m, 2H, Piperidine-H), 2.67 (s, 3H, N-CH3), 2.68 (m, 1H, Piperidine-H), 2.75 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.09 (m, 2H, Ar-H), 7.28 (m, 2H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H);13C NMR spectral data (25.15 MHz, CDCl3): ? 136.1 (C-2), 143.9 (C-4), 149.3 (C-5), 95.5 (C-6), 158.3(C-7), 112.6(C-8), 119.5(C-9), 49.4(C-11), 31.4(C-12), 51.6(C-13), 31.5(C-14), 49.5(C-15), 55.5(C-17), 42.1(C-19), 57.2 (C-20), 136.5(C-21), 129.8 (C-22, C-26), 115(C-23,C-25), 163.4 (C-24);Anal. Calcd for C21H24FN3O2; C, 68.27; H, 6.55; F, 5.14; N, 11.37; O, 8.66; MS (ESI) m/z: 369.1853.
N-(3,4-difluorobenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6e
65 % yield. Mp: 169-171 ºC; 1H-NMR (CDCl3): ? 1.44 (m, 2H, Piperidine-H), 1.67 (m, 2H, Piperidine-H), 2.67 (s, 3H, N-CH3), 2.68 (m, 1H, Piperidine-H), 2.75 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.08 (m, 2H, Ar-H), 7.25 (m, 1H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H); 13C NMR spectral data (25.15 MHz, CDCl3): ? 136.4 (C-2), 144.1 (C-4), 149.2 (C-5), 95.2 (C-6), 158.9(C-7), 112.2(C-8), 119.7(C-9), 49.6(C-11), 31.7C-12), 51.4(C-13), 31.8(C-14), 49.5(C-15), 55.9(C-17), 42.3(C-19), 57.6 (C-20), 140.2(C-21), 129.8 (C-22, C-26), 128.7(C-22), 133.2 (C-23), 133.6(C-24), 129.9, (C-25), 129.2 (C-26); Anal. Calcd for C21H23F2N3O2; C, 65.10; H, 5.98; F, 9.81; N, 10.85; O, 8.26; MS (ESI) m/z: 387.1758.
N-(4-methylbenzene)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6f
61 % yield. Mp: 155-157 ºC; 1H-NMR (CDCl3): ? 0.9 (s, 3H, CH3), 1.42 (m, 2H, Piperidine-H), 1.66 (m, 2H, Piperidine-H), 2.66 (s, 3H, N-CH3), 2.69 (m, 1H, Piperidine-H), 2.75 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.18 (m, 2H, Ar-H), 7.29 (m, 2H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H);13C NMR spectral data (25.15 MHz, CDCl3): ? 136.5 (C-2), 143.7 (C-4), 149.4 (C-5), 95.9 (C-6), 158.4(C-7), 112.5(C-8), 119.8(C-9), 49.1(C-11), 31.2(C-12), 51.9(C-13), 31.1(C-14), 49.3(C-15), 55.2(C-17), 42.7(C-19), 57.4 (C-20), 136.5(C-21), 128.2 (C-22, C-26), 125.1(C-23,C-25), 131.9 (C-24) , 124.1 (C-27); Anal. Calcd for C22H27N3O2; C, 72.30; H, 7.45; N, 11.50; O, 8.76; MS (ESI) m/z: 365.2103.
N-(4-(trifluoromethyl)benzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine6g
70 % yield. Mp: 148-150 ºC; 1H-NMR (CDCl3): ? 1.43 (m, 2H, Piperidine-H), 1.66 (m, 2H, Piperidine-H), 2.65 (s, 3H, N-CH3), 2.69 (m, 1H, Piperidine-H), 2.74 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.18 (m, 2H, Ar-H), 7.29 (m, 2H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H); 13C NMR spectral data (25.15 MHz, CDCl3): ? 136.2 (C-2), 143.5 (C-4), 149.1 (C-5), 95.2 (C-6), 158.7(C-7), 112.3(C-8), 119.4(C-9), 49.3(C-11), 31.4(C-12), 51.7(C-13), 31.5(C-14), 49.4(C-15), 55.3(C-17), 42.2(C-19), 57.6 (C-20), 136.7(C-21), 128.5 (C-22, C-26), 128.3(C-23,C-25), 139.8 (C-24) , 21.3 (C-27); Anal. Calcd for C22H24F3N3O2; C, 63.00; H, 5.77; F, 13.59; N, 10.02; O, 7.63; MS (ESI) m/z: 419.1821.
N-(4-tert-butylbenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine6h
65 % yield. Mp: 168-170 ºC; 1H-NMR (CDCl3): ? 1.33 (s, 9H, CH3), 1.43 (m, 2H, Piperidine-H), 1.66 (m, 2H, Piperidine-H), 2.65 (s, 3H, N-CH3), 2.69 (m, 1H, Piperidine-H), 2.74 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.18 (m, 2H, Ar-H), 7.29 (m, 2H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H);13C NMR spectral data (25.15 MHz, CDCl3): ? 136.5 (C-2), 143.3 (C-4), 149.8 (C-5), 95.3 (C-6), 158.3(C-7), 112.6(C-8), 119.2(C-9), 49.5(C-11), 31.5(C-12), 51.2(C-13), 31.5(C-14), 49.2(C-15), 55.8(C-17), 42.3(C-19), 57.7 (C-20), 136.8(C-21), 128.1 (C-22, C-26), 129.4(C-23,C-25), 144.5 (C-24) , 35.1 (C-27), 33.5(C-28), 29.1(C-29), 14.0(C-30); Anal. Calcd for C25H33N3O2; C, 73.68; H, 8.16; N, 10.31; O, 7.85; MS (ESI) m/z: 407.2573.
N-(4-nitrobenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6i
70 % yield. Mp: 177-179 ºC; 1H-NMR (CDCl3): ? 1.43 (m, 2H, Piperidine-H), 1.66 (m, 2H, Piperidine-H), 2.65 (s, 3H, N-CH3), 2.69 (m, 1H, Piperidine-H), 2.74 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.18 (m, 2H, Ar-H), 7.29 (m, 2H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H);13C NMR spectral data (25.15 MHz, CDCl3): ? 136.3 (C-2), 143.7 (C-4), 149.3 (C-5), 95.5 (C-6), 158.4(C-7), 112.7(C-8), 119.3(C-9), 49.5(C-11), 31.6(C-12), 51.8(C-13), 31.3(C-14), 49.4(C-15), 55.3(C-17), 42.9(C-19), 57.5(C-20), 136.7(C-21), 127.9 (C-22, C-26), 117.3(C-23,C-25), 140.5 (C-24) Anal. Calcd for C21H24N4O4; C, 63.62; H, 6.10; N, 14.13; O, 16.14; MS (ESI) m/z: 396.1798.
N-(2,5-dimethylbenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine6j
55 % yield. Mp: 154-156 ºC; 1H-NMR (CDCl3): 1H-NMR (CDCl3): ? 1.43 (m, 2H, Piperidine-H), 1.66 (m, 2H, Piperidine-H), 2.35 (s, 6H, CH3), 2.65 (s, 3H, N-CH3), 2.69 (m, 1H, Piperidine-H), 2.74 (m, 2H, Piperidine-H), 2.82 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.75 (s, 3H, OCH3), 7.18 (m, 2H, Ar-H), 7.29 (m, 1H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.88 (m, 2H, Benzoxazolyl-H);13C NMR spectral data (25.15 MHz, CDCl3): ? 136.6 (C-2), 143.8 (C-4), 149.2 (C-5), 95.4 (C-6), 158.5(C-7), 112.2(C-8), 119.8(C-9), 49.2(C-11), 31.3(C-12), 51.2(C-13), 31.6(C-14), 49.4(C-15), 55.2(C-17), 42.7(C-19), 53.7(C-20), 131.3(C-21), 129.2 (C-22), 127.8(C-26), 129.1(C-23),128.6(C-25), 139.4 (C-24), 18.7(C-25), 20.9(C-26); Anal. Calcd for C23H29N3O2; C, 72.79; H, 7.70; N, 11.07; O, 8.43; MS (ESI) m/z: 379.226.
N-(2,5-dimethoxybenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6k
67 % yield. Mp: 162-164 ºC; 1H-NMR (CDCl3): 1H-NMR (CDCl3): ? 1.42 (m, 2H, Piperidine-H), 1.64 (m, 2H, Piperidine-H), 2.64 (s, 3H, N-CH3), 2.69 (m, 1H, Piperidine-H), 2.73 (m, 2H, Piperidine-H), 2.83 (m, 2H, Piperidine-H), 3.63 (s, 2H, methylene-H), 3.72 (s, 6H, OCH3), 3.80 (s, 3H, OCH3), 7.18 (m, 2H, Ar-H), 7.29 (m, 1H, Ar-H), 7.42 (m, 1H, Benzoxazolyl-H), 7.89 (m, 2H, Benzoxazolyl-H);13C NMR spectral data (25.15 MHz, CDCl3): ? 136.7 (C-2), 143.5 (C-4), 149.8 (C-5), 95.1 (C-6), 158.2(C-7), 112.7(C-8), 119.5(C-9), 49.3(C-11), 31.9(C-12), 51.4(C-13), 31.3(C-14), 49.3(C-15), 55.4(C-17), 42.2(C-19), 53.7(C-20), 127.7(C-21), 158.1 (C-22), 111.2(C-26), 112.3(C-23),157.7(C-25), 115.3 (C-24), 55.9(C-28), 55.5(C-30); Anal. Calcd for C23H29N3O4; C, 67.13; H, 7.10; N, 10.21; O, 15.55; MS (ESI) m/z: 411.2158.
N-(3,4,5-trimethoxybenzyl)-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine6l
72 % yield. Mp: 169-171 ºC; 1H-NMR (CDCl3): 1H-NMR (CDCl3): ? 1.41 (m, 2H, Pieridine-H), 1.63 (m, 2H, Piperidine-H), 2.65 (s, 3H, N-CH3), 2.68 (m, 1H, Piperidine-H), 2.73 (m, 2H, Piperidine-H), 2.84 (m, 2H, Piperidine-H), 3.61 (s, 2H, methylene-H), 3.75 (s, 9H, OCH3), 3.80 (s, 3H, OCH3), 7.19 (m, 1H, Ar-H), 7.27 (m, 1H, Ar-H), 7.41 (m, 1H, Benzoxazolyl-H), 7.90 (m, 2H, Benzoxazolyl-H);13C NMR spectral data (25.15 MHz, CDCl3): ? 136.6 (C-2), 143.8 (C-4), 149.2 (C-5), 954 (C-6), 158.5(C-7), 112.2(C-8), 119.8(C-9), 49.2(C-11), 31.3(C-12), 51.2(C-13), 31.6(C-14), 49.4(C-15), 55.2(C-17), 42.7(C-19), 53.7(C-20), 142.7(C-21), 107.6 (C-22, C-26), 152.9(C-23,C-25),139.2 (C-24), 56.2(C-28, C32), 60.9(C-30); Anal. Calcd for C24H31N3O5; C, 65.29; H, 7.08; N, 9.52; O, 18.12; MS (ESI) m/z: 441.2264.
Molecular modelling and docking studies
Active site determination for Hsp70 and Hsp90
All the molecular modelling studies were carried out on the Schrodinger modelling suite (Version 15.3). The Heat Shock Proteins (Hsp) 70 and 90 were obtained from Brookhaven Protein Data Bank (RCSB-PDB). Initially the proteins crystals obtained from PDB were analysed and the active sites were determined. Hsp70 (PDB Id: 2XA4) and Hsp90 (PDB Id: 3FGR) were prepared in the protein preparation wizard of the Schrodinger suite for knocking out the bound ligands and cofactors, excess and nonessential water molecules were also removed. The proteins were optimised and missing loops were filled and overlapping residues were corrected, finally both the target proteins were optimised for their conformational energies. These were further subjected to Q-site program to determine the active sites within the protein strucutre.14 The active sites were determined with regions such as hydrophobic and polar-regions as presented in Figure 2.
Molecular docking studies
The compounds designed were subjected to energy minimisation by Ligprep module of the software. Glide module was employed for the molecular docking; compounds were docked in the grid designed in the module. The docking was carried out with flexible ligands and the protein with the simulations made on the OPLS5.0 force field. All the results were based on 10 conformations for each molecule in the standard precision (SP) mode. The crystal structure Hsp70 (PDB Id: 2XA4) and Hsp90 (PDB Id: 3FGR) were both employed as the receptor with compounds 6a-l as ligands.14) The results obtained from the simulation were obtained in the form of dock score and Glide energy, these values represents the minimum energies. Interactions between the ligand and residues were presented in the form of H-bond, van der Waals forces and the pi bonds. The results in the form of 3D and 2D representation were obtained for simplified understanding and presented as Figure 2 and Figure 3

Figure 2: a) Active site determination in the Hsp70 (PDB Id: 2XA4); b) Active site determination in the Hsp90 (PDB Id: 3FGR)
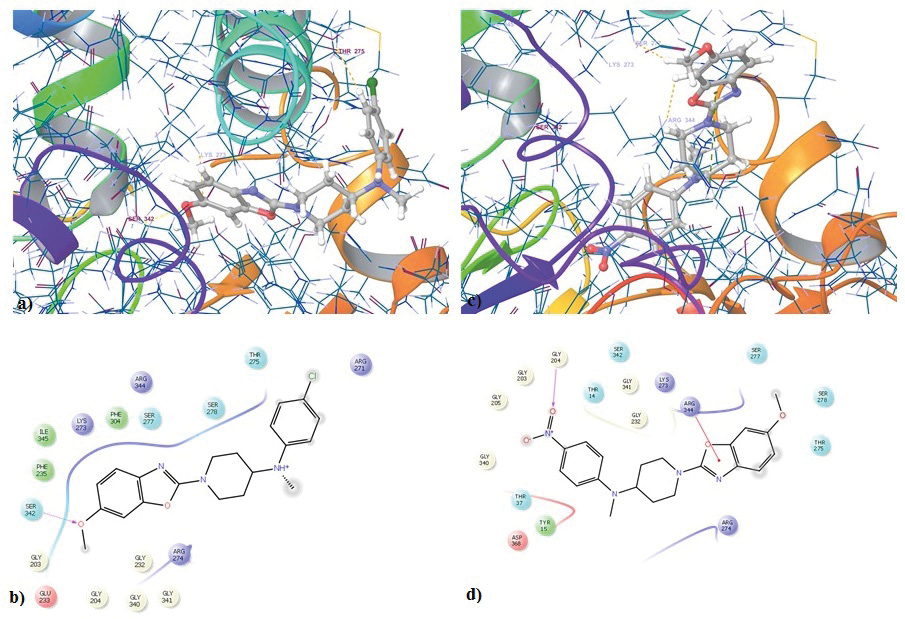

Figure 3: Molecular docking study on compounds from the series; a,b) 3D and 2D representation of compound 6bonHsp70. (PDB Id: 2XA4); c,d) 3D and 2D representation of compound 6ion Hsp70 (PDB Id: 2XA4).
IC50 for both the compounds are explained in Figure 4A for 6band 6i.
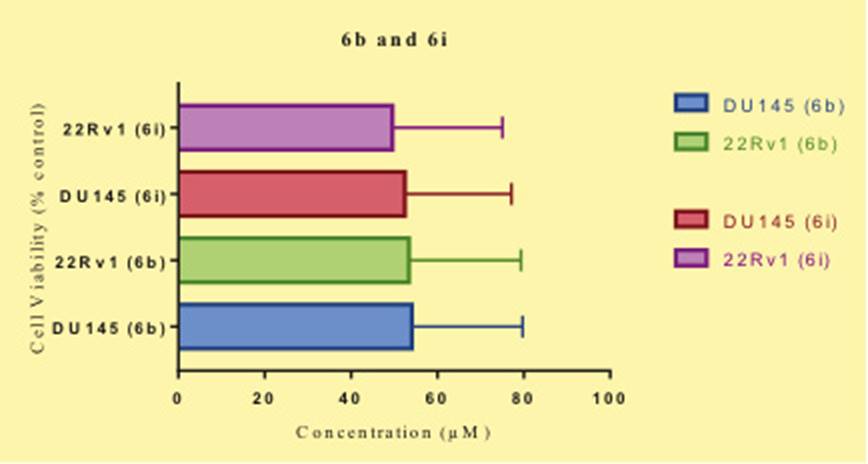
Figure 4: A) Molecular docking study on compounds from the series; a, b) 3D and 2D representation of compound 6b onHsp90 (PDB Id: 3FGR); c, d) 3D and 2D representation of compound 6i on Hsp90 (PDB Id: 3FGR). B) Compound 6b and 6i inhibited overall cell viability (% relative to control) DU-145 and 22Rv1 (Cell lines for Prostate cancer). The cell viability was determined by plotting CCK-8 assay with increasing concentration of compoundsfor 24 hours.
Table 1: Antitumor activity of some novel hybrids of benzoxazole,the table represents concentration as a result of 50% loss of viable cells with respect to untreated cells (IC50). The results were defined from dose-response curves for two different cell lines DU-145 and 22Rv1. It also presents the glide score and energies for different derivatives in front respective derivatives.
| Sr. no. | Compd no. | IC50 (µM) | Dock score | ||||
|---|---|---|---|---|---|---|---|
| DU145 | 22Rv1 | Hsp 70 | Hsp 90 | ||||
| Dock Score | Glide Energy | Dock Score | Glide Energy | ||||
| 1 | 6a | 28.91 | 27.51 | -8.0251 | -46.3115 | -5.07476 | -34.435 |
| 2 | 6b | 20.10 | 19.41 | -8.12051 | -46.1657 | -5.08201 | -49.2727 |
| 3 | 6c | >100 | 48.12 | -7.76383 | -48.4989 | -4.41685 | -43.5636 |
| 4 | 6d | 47.45 | 44.94 | -7.91281 | -46.2466 | -5.09304 | -39.2727 |
| 5 | 6e | 29.35 | 45.21 | -8.02916 | -49.2831 | -5.34989 | -46.6876 |
| 6 | 6f | >100 | 43.33 | -8.02665 | -46.5599 | -3.7217 | -35.7962 |
| 7 | 6g | 45.24 | 35.55 | -7.85481 | -48.8817 | -3.39334 | -35.8336 |
| 8 | 6h | 41.42 | 39.92 | -7.51404 | -46.4911 | -3.51714 | -38.4987 |
| 9 | 6i | 20.5 | 19.00 | -8.84901 | -49.7121 | -5.34706 | -46.6326 |
| 10 | 6j | >100 | 43.36 | -7.70333 | -45.1409 | -4.43773 | -32.1075 |
| 11 | 6k | >100 | 44.33 | -7.32014 | -46.7713 | -4.98797 | -37.4438 |
| 12 | 6l | 47.10 | 44.89 | -7.63666 | -49.3863 | -3.6077 | -39.2424 |
| 13 | Std Enzalutamide | 28.22 | 31.72 | -7.344 | -46.7819 | -5.09304 | -39.2727 |
In vitro screening
Cell Culture
The cell lines DU-145 and 22Rv1 were obtained from the ATCC, USA; DU145, 22Rv1 and RWPE-1 werepreserved at -80 ºC according to the standard protocol. These were further cultured in Dulbecco’s Eagle medium modified and supplemented, it contains fetal Bovine Serum (10%) along with penicillin-100 U/mL and streptomycin-0.1 mg/mL as per the requirement. Before use these cell lines were kept for incubation at 37 ºC with controlled humidity and 5 % carbon dioxide.
Oncotest assay method for anitproliferative activity15),(16
Oncotest’s monolayer assay was developed for testing the antiproliferative activity of newly developed compounds was found to the most accurate and suitable method for evaluation of these compounds. This method was found to be highly accurate and specific for testing of compounds on prostate cancer cell lines. The widely used anticancer drug enzalutamide was used as standard drug on both the cell lines along with other test compounds. This assay is based on the propedium iodide method, the test compounds are observed after four days of treatment. All the derivative 6a-l were tested for ten dilutions with half log increasing to the 100 1M. The readings were taken in triplicates and observations were presented by nonlinear regression analysis as IC50 in Table 1 forming the results. The detailed experimental protocol is provided with the supplementary material available online.
Antitumor Activity by CCK-8 Assay17
The CCK-8 assay stands for Cell Counting Kit 8 (CCK 8) which was developed by Dojindo Corporation in Japan. It is a ready to use kit to determine the extent of proliferation by both cell lines DU-145 and 22Rv1 the selected compounds 6b and 6i. (Figure 4B). Three replicates for each of the compound was carried out and the cell growth inhibition was calculated as the following formula (Formula 1);
Surface Plasmon Resonance (SPR) Analysis18
All the SPR readings were obtained from optical biosensor Biacore T100 (GE Company) reported elsewhere. The human HSP70 (ADI-ESP-550-F [200 µg], Enzo Life Sciences)was immobilized on a CM5 sensor chip (GE) using 10 mmol/PBS at pH 5.0. The binding experiments were carried out at 25 ºC using a flow rate of 30 mL/L at 90 s of monitoring for association and 120 s of monitoring fordissociation of the ligand and substrate. All studies were performed witheight dilutions as 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, and10 mM, with all of these prepared with 5% DMSO in PBS along with it 5% DMSO in PBS was used as a running buffer andblank control. The datawasdelivered using equilibrium dissociation constant (Kdvalue). This was calculated following equation as:
Response = Conc x Rmax / [conc + Kd] + offset
RESULTS AND DISCUSSION
Prostate cancer has been treated with the help of chemotherapy and surgery, but rise in incidences of drug resistance in prostate carcinoma have inclined the treatment more towards surgery. This lead to need for newer and better drug molecules to treat the disease and hence forth discovery of newer targets. Hsp70 and Hsp90 has been studied for their role in the prostate carcinoma, it was found that there is an elevation and elevated expression of these proteins in the patients suffering from cancer. Further it was established that upregulation of these to heat shock proteins plays a significant role in cancer progression and hence are good targets for treatment of prostate cancer. Herein we have describe design and development of few novel inhibitors of Hsp70 and Hsp90 for their application towards treatment of prostate cancer. The design of new molecules is based on the structural features like the fused aromatic nucleus, cyclic aliphatic group and halogen substitutions on the aromatic ring. These features are found to me common in many anticancer agents with use in treatment of prostate cancer. On the basis of hybrid approach we designed a series of derivatives with a fused aromatic nucleus the benzoxazole, cyclic aliphatic group the piperidine linker and aromatic ring with various substitutions.
The synthesis was carried out in four steps, first step involved substitution of piperidine ring 2 on the 2-chloro benzoxazole 1 leading to formation of the tert-butyl 1-(6-chlorobenzo[d]oxazol-2-yl)piperidin-4-yl(methyl)carbamate 3, this was furthertreated with sodium methoxide in methanol to obtain tert-butyl 1-(6-methoxybenzo[d]oxazol-2-yl)piperidin-4-yl(methyl) carbamate 4. This was deprotected by removing Boc to yield1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 5. In the final step various phenyl chlorides were substituted to yield corresponding derivatives as N-substituted benzyl-1-(6-methoxybenzo[d]oxazol-2-yl)-N-methylpiperidin-4-amine 6a-l. These compounds were characterisedfor their structural confirmation and molecular structures were established.
To study the biological efficacy of these compounds oncotest assay method was performed on two cell lines DU145 and 22Rv1. The results obtained from this assay shows that two compounds 6b and 6i have good IC50 values (Table 1). Compound 6b had IC50 of 20.10 and 19.41 for the DU145 and 22Rv1 respectively, while 6i had IC50 of 20.5 and 19.00 for DU145 and 22Rv1 respectively, this is quit satisfactory compared to the standard drug enzalutamide. In the subsequentevaluation these compounds were studied on the basis of CCK-8 assay for the two cell lines DU145 and 22Rv1 and cell viability was studied. It was found that compound 6i inhibits the 22Rv1 more efficiently ten Du145 whereas 6b has more inhibitory activity on the 22Rv1 (Fig 5). In order to understand the mechanism of action of these two molecules molecular docking studies were carried out. Two different receptors were obtained from the protein data bank, Hsp70 (PDB ID: 2XA4) and Hsp90 (PDB ID: 3FGR). Initially these two proteins were subjected to protein preparation leading to filling of missing loops and resolving the overlapping residues. After optimising these protein on the Schrodinger protein preparation wizard these were subjected to Qsite finder. Qsite determines the active site within the receptor which is important for determining the residues involved in interaction with the ligands (Figure 2). In case of Hsp70 it was found that residues Leu40, Val186, Asp93, Trp162, Phe138, Ser52, Leu103, Asn51 and Gly100 form the active site and consist of an cavity within the receptor. In Hsp90 the active site was formed of Ser342, Phe235, Ile345, Lys273, Ser277, Arg274, Ser278, Glu233 and Arg344.
The molecular docking was carried out for the two compounds 6b and 6i in these active sites of Hsp70 and Hsp90 with the help of Glide module of Schrodinger software version 9.5. The compounds were docked on both the receptors and scores were obtained for their binding glide energy and dock score. The results are shown in Table 1, it can be seen that both compounds have performed well in the docking studies. Compound 6b has good dock score of -8.12 and Glide score of -46.16 for Hsp70, for Hsp90 it had dock score of -5.08 and Glide energy of -49.27. Compound 6i has good dock score of -8.84 and Glide score of -49.71 for Hsp70, for Hsp90 it had dock score of -5.34 and Glide energy of -46.63. Figure 3 shows the tree dimensional and two dimensional images of the compounds 6b and 6i interacting with Hsp70. It is clearly observed that the compound6b forms hydrogen bond with the amine group of the linker and Asp93, two pi bond interactions are observed between the Phe138 and Trp162 with the benzoxazole nucleus. Compound 6iinteracts with the receptor forming hydrogen bonds with Asn51 and Asp93, pi bond between Phe138 and Trp162, hydrogen and salt bridge with the Lys58. Figure 4 shows the tree dimensional and two dimensional images of the compounds 6b and 6i interacting with Hsp970. It is observed that 6b forms an hydrogen bond with Ser342 and 6i forms an pi bond with the Arg344 These results suggest that compound 6b and 6i have high affinity for the Hsp70 compared to Hsp90.
In order to further investigate the efficacy of compound 6b and 6i we carried out the drug binding assay based on the surface plasmone resonance (SPR) principle on the Hsp70. The experiment was carried out on the human Hsp70 and the association and disassociation of the ligand with the substrate was determined (Table 2). The dissociation constant (Kd) was calculated and it was found that compound 6b had Kd = 2.030 x 10-6and 6i had Kd = 2.421x 10-6compared to the standard with Kd = 2.066 x 10-6. This indicates that compound 6b and 6i are having comparable affinity towards the Hsp90.
CONCLUSION
Present study provides with two novel molecules 6b and 6i, these have good inhibitory activity against the Hsp70 and Hsp90, but it is more specific for the inhibition of the Hsp70. The results obtained and the observation make it clear that these compounds have good in vitro activity against prostate cancer. These compounds can be further evaluated for their in vivo activity against the prostate cancer and developed into lead molecules.