Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Archivos de Zootecnia
versión On-line ISSN 1885-4494versión impresa ISSN 0004-0592
Arch. zootec. vol.60 no.231 Córdoba sep. 2011
https://dx.doi.org/10.4321/S0004-05922011000300049
Genetic diversity of Albanian goat breeds based on microsatellite markers
Diversidad genética de razas caprinas de Albania, basada en marcadores microsatélites
Hoda, A.1*, Hyka, G.1A, Dunner, S.2, Obexer-Ruff, G.3 and Econogene Consortium4
1Department of Animal Production. Agricultural University of Tirana. AUT. Albania. *hodanila@yahoo.com; Agentianhyka@yahoo.com
2Department of Animal Production. Facultad de Veterinaria. Universidad Complutense. Madrid. España. dunner@vet.ucm.es
3Institute of Genetics. Veterinary Faculty. University of Berne. Berne. Switzerland. gabriela.obexerruff@itz.unibe.ch
4Members of the Econogene Consortium: Abo-Shehada Mahamoud, Ajmone Marsan Paolo, Al Tarrayrah Jamil, Angiolillo Antonella, Baret Philippe, Baumung Roswitha, Beja-Pereira Albano, Bertaglia Marco, Bordonaro Salvatore, Bruford Mike, Caloz Régis, Canali Gabriele, Canon Javier, Cappuccio Irene, Carta Antonello, Cicogna Mario, Crepaldi Paola, Dalamitra Stella, Daniela Krugmann, Dobi Petrit, Dominik Popielarczyk, Dunner Susana, D'Urso Giuseppe, El-Barody M. A. A, England Phillip, Erhardt Georg, Ertugrul Okan, Eva- Maria Prinzenberg, Eveline Ibeagha-Awemu, Ewa Strzelec, Fadlaoui Aziz, Fornarelli Francesca, Garcia David, Georgoudis Andreas, Gesine Lühken, Giovenzana Stefano, Gutscher Katja, Hewitt Godfrey, Hoda Anila, Horst Brandt, Istvan Anton, Juma Gabriela, Joost Stéphane, Jones Sam, Karetsou Katerina, Kliambas Georgios, Koban Evren, Kutita Olga, Lazlo Fesus, Lenstra Johannes A, Ligda Christina, Lipsky Shirin, Luikart Gordon, Marie-Louise Glowatzki, Marilli Marta, Marletta Donata, Milanesi Elisabetta, Negrini Riccardo, Nijman Isaäc J, Obexer-Ruff Gabriela, Papachristoforou Christos, Pariset Lorraine, Pellecchia Marco, Peter Christina, Perez Trinidad, Pietrolà Emilio, Pilla Fabio, Roman Niznikowski, Roosen Jutta, Scarpa Riccardo, Sechi Tiziana, Taberlet Pierre, Taylor Martin, Togan Inci, Trommetter Michel, Valentini Alessio, Van Cann, Lisette M, Vlaic Augustin, Wiskin Louise, Zundel Stéphanie. http://www.econogene.eu
This work has been supported by theEconogene project, funded by the EuropeanUnion (project QLK5- CT2001-02461).
SUMMARY
The domestic goat is one of the most important livestock species in mountainous area of Albania. In this study thirty microsatellite markers in 183 unrelated individuals from 6 local goat breeds are analyzed. Twenty nine markers had five or more alleles. All loci were polymorphic and a total of 331 alleles were detected. The average number of alleles per locus was 11.03. Within breeds, the mean number of alleles ranged from 7.8 to 8. Mean expected heterozygosity (He) ranged from 0.712 to 0.758. Allelic richness varied from 7.61 to 8.19. Inbreeding for all population is rather high FIS= 0.093, ranging from 0.075 to 0.103. The mean FST (˜ 0.02) demonstrated that 98% of total genetic variation is due to genetic differentiation within each population.
Key words: Albanian local breeds.
RESUMEN
La cabra doméstica es una de las especies ganaderas más importantes en la zonas montañosas de Albania. En este trabajo se analizaron 30 marcadores microsatélites procedentes de 183 individuos no relacionados, pertenecientes a 6 razas locales de cabras. Veintinueve marcadores mostraron 5 o más alelos. Todos los loci fueron polimórficos habiéndose detectado un total de 331 alelos. El número medio de alelos por locus fue 11,03. Dentro de las razas, el número medio de alelos osciló entre 7,8 y 8. La heterocigosidad media esperada (He) osciló entre 0,712 y 0,758. La riqueza alélica varió de 7,61 a 8,19. La consaguinidad para toda la poblacón es más bien alta FIS= 0,093 con un rango de 0,075 a 0,103, la FST media (˜ 0,02) demostró que el 98% de la variación genética total se debe a la diferenciación genética dentro de cada población.
Palabras clave: Razas locales de Albania.
Introduction
The domestic goat is one of the most important livestock species in mountainous area of Albania. Goat breeds are defined mainly by the geographic location, morphological characteristics and production performance.
Polymorphic DNA markers are very useful in assessment of genetic diversity within and between breeds. Microsatellites are widely used as genetic markers for the analysis of genetic variability within and between breeds due to their high number, distribution throughout the genome and the efficacy of genotyping.
There are several studies on genetic diversity of goats, based on microsatellite markers, such as Swiss breeds (Saitbekova et al., 1999), Chinese indigenous populations (Liet al.,2002, Liet al.,2004), Indian domestic goats (Rout et al., 2008), goats from Europe (including also the breeds represented here) and middle east (Cañon et al., 2006). In this study, carried out in the frame of econogene project, we intend to determine the levels of genetic variation and relationships among six local goat breeds, representing diverse morphological characteristics. This is the first effort to characterize the Albanian local goat breeds through DNA markers.
Material and methods
SAMPLE COLLECTION AND MICROSATELLITE MARKERS
A total of 183 randomly sampled animals representing 6 different Albanian goat breeds were analyzed. For each breed a maximum of three unrelated individuals (two females and one male) per flock were sampled, based on the information provided by the farmer. Sampling was carried out from an average of 11 flocks per breed. The breeds are marginally farmed and locally distributed. Thirty microsatellite markers according to MoDAD (ISAG/FAO standing committee) were used to genotype the sampled animals using procedures detailed elsewhere (Cañon et al., 2006).
STATISTICAL ANALYSES
Allele frequencies and tests of genotype frequencies for deviation from Hardy-Weinberg Equilibrium (HWE) were carried out using exact tests of the Genepop V.1.2 program (Rousset et al., 2008). Genetix version 4.05.2 (Belkhir et al., 2001) (http://www.univ-montp2.fr//~genetix/genetix/genetix/htm) was used for the calculation of observed heterozygosity (Ho), mean unbiased estimates of gene diversity (He) and the F-statistics (Wright, 1978, Wright, 1965), FIS, FIT and FST including their significance level by a permutation test. The program FSTat (Goudet et al., 1995) (http://jhered.oxfordjournals.org/cgi/content/citation/86/6/485) was used for the calculation of corrected allele diversity (allelic richness).
Nei genetic distance (Nei, 1972) was calculated and used for the construction of UPGMA consensus tree (Saitou and Nei, 1987) with phylip package (http:// evolution.genetics.washington.edu/phylip.html). Bootstrap (1000 replicates) resampling was performed to test the robustness of the dendrogram topology. Nei genetic distance calculated from the allele data was plotted as PCA using GenAlEx program (Peakall and Smouse, 2006).
Assignment of individual to their reference population was evaluated using GeneClass v.1.0.02 (http://www.ensam.inra.fr/URLB/geneclass/geneclass.html) (Cournet et al., 1999). For allbreeds was carried out a direct assignment of individuals and a exclusion analysis based on 10 000 simulated individuals (Cournet et al., 1999). The methods based on allele frequencies (Paetkau et al., 1995) as well as bayesian approach (Rannala and Mountain, 1997) were used. The calculations were carried out using always a leave one out procedure.
The program structure (Pritchard et al., 2000) is used for the analysis of population methods based on allele frequencies structure by a clustering analysis based in (Paetkau et al., 1995) as well as bayesian bayesian model. The program uses markov approach (Rannala and Mountain, 1997) chain monte carlo method the program was run under theadmixture model, with burning period of 100 000 iterations and period of data collection of 100 000 iterations. The samples were analyzed withK from 2 to 7, applying 3 running.
Results and discussion
MICROSATELLITE LOCI
All the markers were polymorphic. The total number of alleles and allele size for each locus are presented in table I. In total 331 alleles were observed over all loci across the 183 individuals. The mean number of alleles per locus was 11 and varied from 4 (MAF209) to 30 (BM6444). Out of 30 markers, 29 showed 5 or more alleles. The total number of alleles per locus in the present study ranged from 5 to 30, except of MAF 209 which had 4 alleles. Observed Ho per locus ranged from 0.41 (InraBern185) to 0.8 (SRCRSP23), with an average of 0.67. A total of 43 breed specific alleles were detected at 25 loci, but only 5 of these alleles had frequencies ≥5%. Departures from Hardy-Weinberg proportions were revealed in 43 of the 180 locus-population comparisons (p<0.05). The loci DRBP1, P19 deviated in all populations and BM6444 deviated in five populations. These markers were excluded from the genetic differentiation analyses.
GENETIC VARIATION WITHIN AND AMONG BREEDS
In table II the measures of genetic variability of 6 Albanian goat breeds are presented. The most diverse breeds were Muzhake and Hasi with the highest total number of alleles (TNA) of 240 and the highest mean number of alleles (MNA) of 8. Capore and Dukati have the lowest TNA of 233 and the lowest MNA of 7.8. Expected heterozygosity was lowest in Mati of 0.71 and the highest in Muzhake of 0.76. Allelic richness (AR) ranged from 7.6 in Capore to 7.8 in Muzhake with an average of 7.7 alleles per breed. The mean expected He ranged from 0.71 in Mati to 0.76 in Muzhake. Observed Ho varied from 0.64 in Mati to 0.69 in Muzhake. Mean values of He and Ho, overall loci and breeds were 0.74 and 0.67 respectively. The mean number of alleles per locus (MNA) varied from 7. 8 in Capore and Dukati to 8 in Hasi and Muzhake. FIS values ranged from 0.076 in Dukati to 0.105 in Capore. table II shows the number of loci deviating significantly (p<0.05) from HWE, per each breed.
All breeds displayed significant deviation from HWE in more than 6 loci. The positive FIS values showed heterozygotes deficiency within breeds. This deficit might be because of inbreeding and wahlund effect. Inbreeding values obtained for all the breeds varied from 0.076 in Dukati to 0.105 in Capore with a mean value of 0.093. Cañon et al., 2006 reported an average FIS value of 0.10 for the 45 goat breeds from Europe (including also 6 Albanian local breeds) and Middle East analyzed with the same set of microsatellite markers. These high inbreeding values could be due to their small population size, small number of breeding males and their limited geographical area of dispersion (Dobi et al., 2006). Another possible reason for the high values of inbreeding may be the wahlund effect, because the sampling is carried out in 11 flocks per breed with a probable genetic differences among them.
GENETIC DIFFERENTIATION
The average genetic differentiation between all breeds (FST value) was 0.02, significantly different from zero (p<0.001), which means that about 2% of the total genetic variation was explained by population differences and 98% correspond to the differences among individuals within each breed.
Genetic distance are small and ranged from 0.071 to 0.153. Small genetic distances have also been found among Subsaharian Africa goat breeds (Muema, 2009), or among Indian goat breeds (Shadma 2006). Corresponding to these small genetic distances high gene flow between breeds is expected under an island model and values found ranged from 7.5 (Liqenasi-Dukati) to 31.8 (Capore-Muzhake).
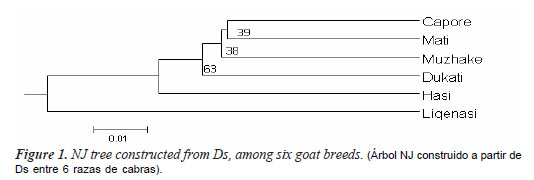
Nei's distance (Nei, 1972) (DS) between each pair of populations (table III) was generated based on allele frequencies and used to build a dendrogram with the NJ algorithm (figure 1). The number at the nodes are values for 1000 bootstrap resampling of the typed loci. The NJ tree revealed a cluster of Capore and Mati, although bootstrap value was low.
The principal component analysis (PCA) based on allele frequencies is presented in figure II. It is clear that Capore, Muzhake and Dukati form one group. Again, Liqenasi seems quite separate in a single quadrate. The first component clearly discriminate Liqenasi from the rest, while the second component discriminate Hasi, Mati, Dukati and the rest three breeds.
The genetic analysis of 6 Albanian local goat breeds with 30 microsatellite markers showed high gene diversity. The high number of alleles for each locus and the high gene diversity for each breed showed the appropriateness of the markers to analyze diversity in Albanian local goat breeds.
Both NJ tree and PCA analysis showed that Liqenasi breed is distinct from the other breeds. This breed is located in an isolated area in South east of Albania where population has the Macedonian nationality, therefore they prefer to have limited relationship with other areas of Albania (Dobi et al, 2006). FST among the local goat breeds in this study is 2%, that is very low compared to the values of 6.9% reported by Cañon et al., 2006, the 14% for Asian goats (Barker et al., 2001), the 17% for Swiss goat breeds (Saitbekova et al, 1999), the 10.5% for Chinese goat breeds (Li et al, 2002), 5.4% West African Dwarf goat (Mujibi, 2005), or even the 5% found for indigenous goats of Sub-Saharan Africa (Muema, 2009). All pairwise FST values were significant (p<0.01). The lack of herd book, until nowadays, probably has facilitate the admixture of the breeds contributing to a high level of gene flow between the breeds reducing, as a consequence, the level of genetic differentiation. In practice, the reproducing males are selected by the farmers, who try to avoid the use of males from their own flock, but usually buy them in the farm animal market, or from neighbor farms without any information or control of their origin, resulting in mating without parentage control.
The assignment (direct) and exclusion (simulation) of individuals to their reference population (table IV) is carried out by frequency, and bayessian methods. The confidence level was 99%. The Bayes approach performed better for assigning animals to their breed. Muzhake was the breed with a lower rate of animals correctly assigned, and Liqenasi, the breed with the higher rate of correctly assigned animals, being also this breed which showed the higher rate of excluded animals (25.8). The relatively low percentage of correctly assigned individuals is in concordance with the small genetic distance between breeds and with the low genetic differentiation (FST˜ 2).
After running the program structure, with the K ranging from 2 to 7 we found that the highest value ln Pr(X|K) was obtained for K= 2-3, but the variance is higher at K= 3. In table V the proportions of membership of each breed in inferred clusters for K= 2 and K= 3 are presented. In both cases Liqenasi breed is more differentiated, 60% (K= 2) and 73% (K= 3) of individuals are assigned in a single cluster. The other breeds show a high level of admixture.
The small ruminant farming is one of the main production activities in hill and mountainous area of Albania and consists in small farms with limited number of effectives managed on extensive or semiextensive system with natural mating, which provide an important source of meat and milk mainly for family consumption. Product marketing and processing is limited and difficult due to the low rural socio economic level, poor infrastructure and investments. Goat well adapted to the harsh mountainous environment, poor pasture, and resistant to diseases, use natural pastures wit spontaneous flora resources (concentrated feed is used only before and after weaning). The number of goats is decreasing rapidly in the last years the main reason being possibly the movement of rural population towards urban areas, youth emigration abroad, due to the low economic development of these regions. There is no breeding program for these goat breeds, and the genetic characterization of these breeds carried in the present study, may be used to start a breeding strategy and a policy in order to conserve valuable breeds.
Acknowledgements
The content of the publication does notrepresent necessarily the views of the commission or its services.
Bibliography
Barker, J.S.F., Tan, S.G., Moore, S.S., Mukherjee, T.K., Matherson, J.L. and Selvaraj, O.S. 2001. Genetic variation within and relationship among populations of Asian goats (Capra hircus). J. Anim. Breed. Genet., 118: 213-233. [ Links ]
Belkhir, K., Borsa, P., Chikhi, L., Raufaste, N. et Bonhomme, F. 2001. Genetix, logiciel sous windows TM pour la génétique des populations. Laboratoire génome, populations, interactions. CNRS UPR 9060. Université de Montpellier II. Montpellier. France. http://www.genetix.univmontp2.fr/genetix/intro.htm. (10/12/09). [ Links ]
Cañon, J., Garcia, D., Garcia-Atance, M.A., Obexer-Ruff, G., Lenstra, J.A., Ajmone-Marsan, P., Dunner, S. and Econogene Consortium. 2006. Geographical partitioning of goat diversity in Europe and the Middle East. Anim. Genet., 37: 327-334. [ Links ]
Cornuet, J.M., Piry, S., Luikart, G., Estoup, A. and Solignac, M. 1999. New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics, 153: 1989-2000. [ Links ]
Dobi, P., Hoda, A., Sallaku, E. and Kolaneci, V. (Eds.). 2006. Racat autoktone të bagëtive të imta. 1ST. Tirana 2000. Albania. 125 pp. [ Links ]
Goudet, J. 1995. A computer program to calculate F-statistics. FSTAT (version 1.2). http://jhered.oxfordjournals.org/cgi/content/citation/86/6/485. J. Hered., 86: 485-486. [ Links ]
Li, M.H., Zhao, S.H., Bian C., Wang, H.S., Wei, H., Liu, B., Yu, M., Fan, B., Chen, S.L., Zhu, M.J., Li, S.J., Xiong, T.A. and Li, K. 2002. Genetic relationships among twelve Chinese indigenous goat populations based on microsatellite analysis. Genet. Sel. Evol., 34: 729-44. [ Links ]
Li, S.L. and Valenti, A. 2004. Genetic diversity of Chinese indigenous goat breeds based on microsatellite markers. J. Anim. Breed. Genet., 121: 350-55. [ Links ]
Muema, E.K., Wakhungu, J.W., Hanotte, O. and Jianlin H. 2009. Genetic diversity and relationship of indigenous goats of Subsaharan Africa using microsatellite DNA markers. Livestock Res. Rur. Develop. Volume 21. Article 28. http://www.lrrd.org/lrrd21/2/muem21028.htm [ Links ]
Mujibi, N.F. 2005. Genetic characterization of West African Dwarf goats using microsatellite markers. Msc thesis submitted to Department of Biochemistry and Biotechnology. Kenyatta University. Nairobi. Kenya. [ Links ]
Nei, M. 1972. Genetic distances between populations. Am. Nat., 106: 283-292. [ Links ]
Paetkau, D., Calvert, W., Stirling, I. and Strobeck, C. 1995. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol., 4: 347-354. [ Links ]
Peakall, R and Smouse, P.E. 2006. Population genetic software for teaching and research. GenAlEx 6: genetic analysis in Excel. Mol. Ecol. Notes., 6: 288-295. [ Links ]
Prichard, J.K., Stephens, M. and Donnelly, P. 2000. Inference of population structure using multilocus genotype data. Genetics, 155: 945-959. [ Links ]
Rannala, B. and Mountain, J.L. 1997. Detecting immigration by using multilocus genotypes. USA. Proc. Natl. Acad. Sci., 98: 9197-9201. [ Links ]
Raymond, M. and Rousset, F. 2001. Genepop a population genetics software for exact tests and ecumenicism. http://jhered.oxfordjournals.org/cgi/reprint/86/3/248. (10/12/09). [ Links ]
Rout, P.K., Joshi, M.B., Mandal, A., Laloe, D., Singh, L. and Thangaraj, K. 2008. Microsatellite based phylogeny of Indian domestic goats. Published online 28/01/08. doi: 10.1186/1471-2156-9-11. BMC Genetics., 9: 11. [ Links ]
Saitbekova, N., Gaillard, C., Obexer-Ruff, G. and Dolf, G. 1999. Genetic diversity in Swiss goat breeds based on microsatellite analysis. Anim. Genet., 30: 36-41. [ Links ]
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol., 4: 406-425. [ Links ]
Shadma Fatima. 2006. Study of genetic variability among Gohilwadi, Surti and Zalawadi Goats using microsatellite analysis. Msc thesis submitted to Department of Animal Genetics and Breeding. College of Veterinary Science and Animal Husbandry Anand Agricultural University Anand. India. 120 pp. [ Links ]
Wright, S. 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution, 19: 395-420. [ Links ]
Wright, S. 1978. Evolution and the genetics of populations: variability within and among nature populations. Chicago University Press. Chicago II. USA. 4 pp. [ Links ]
Recibido: 16-10-09
Aceptado: 24-2-10