Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Archivos de Zootecnia
versión On-line ISSN 1885-4494versión impresa ISSN 0004-0592
Arch. zootec. vol.63 no.241 Córdoba mar. 2014
https://dx.doi.org/10.4321/S0004-05922014000100005
Using pedigree analysis to monitor the local Piau pig breed conservation program
Utilização de pedigree para o monitoramento do programa de conservação da raça local de suíno Piau
Veroneze, R.1*; Lopes, P.S.1; Guimarães, S.E.F.1; Guimarães, J.D.2; Costa, E.V.1; Faria, V.R.1 and Costa, K.A.3
1Departamento de Zootecnia. Universidade Federal de Viçosa. Viçosa, MG. Brazil. *renata.veroneze@ufv.br
2Departamento de Medicina Veterinária. Universidade Federal de Viçosa. Viçosa, MG. Brazil
3Departamento de Biologia. Universidade Federal de Viçosa. Viçosa, MG. Brazil
We thank the CNPq (National Council of Research and Development) and FAPEMIG (Minas Gerais State Research Foundation) for financial support.
SUMMARY
The local Brazilian pig breed Piau is a lard-type pig that has undergone a breeding and selection process devised by the veterinarian Antonio Teixeira Vianna in 1939. The objective of the program was to develop a dual purpose animal, that is an animal suitable for the production of both fat and meat. The main characteristics of the Piau breed are rusticity, adaptability and high disease resistance. The conservation status of the breed is threatened, and in 1998, the Pig Breeding Farm at Universidade Federal de Viçosa (UFV, Viçosa, MG, Brazil) started a Piau herd for the purposes of conservation and use in genetic studies. The objectives of the present study were to describe the Piau genetic conservation program of the Universidade Federal de Viçosa (UFV) and to evaluate a range of population parameters to generate information for monitoring and improving the program. Population genetic structure analysis was performed using a pedigree file of 1349 animals and ENDOG v 4.8 software. The Piau conservation program has data and a collection of pedigree registers going back to the introduction of the first animals to the Pig Breeding Farm. The program used mating control, which is based on the mating of animals with the lowest pedigree-based relationships and on the maintenance of all the population's founding families. The effective numbers of founders and ancestors in the studied Piau population are 9 and 8 animals, respectively. The average inbreeding coefficient (F), the average relatedness coefficient (AR) and the effective population size are 6.55 %, 19.74 % and 18.59, respectively. The findings support an increase in the effective population size of this population through the introduction of animals from other populations, as the small number of founders makes inbreeding control difficult.
Key words: Effective population size. Genetic resource. Inbreeding. Population structure.
RESUMO
A raça local brasileira Piau é um suíno tipo banha a qual passou por um processo de seleção realizado pelo médico veterinário Antonio Teixeira Vianna em 1939, com o intuito de desenvolver um animal de dupla aptidão, em outras palavras que fosse adequado para a produção de carne e gordura. As principais características da raça são rusticidade, adaptabilidade e resistência a doenças. O status de conservação da raça é ameaçado e em 1998 a Granja de Melhoramento de Suínos da Universidade Federal de Viçosa (UFV, Viçosa, MG, Brazil) iniciou uma criação de suínos da raça Piau com o objetivo de conservála e utilizá-la em estudos genéticos. O objetivo do presente estudo é descrever o programa de conservação da raça Piau da UFV e avaliar uma série de parâmetros populacionais, a fim de gerar informações que possibilitem o monitoramento e a melhoria do programa. Para a análise da estrutura populacional foi utilizado arquivo de pedigree com 1349 indivíduos por meio do software ENDOG v. 4.8. O programa de conservação da raça possui boa estrutura com registro de dados zootécnicos e pedigree desde a introdução dos primeiros animais na granja de melhoramento de suínos, juntamente com o controle de acasalamentos, o qual é baseado no acasalamento de animais com menor coancestralidade e na manutenção das famílias fundadoras. A população de Piau avaliada possui número efetivo de fundadores e ancestrais iguais a 9 e 8, respectivamente. O coeficiente de endogamia médio, coeficiente de relação médio e tamanho efetivo da população são 6,55 %, 19,74 % e 18,59, respectivamente. De acordo com os parâmetros avaliados é recomendado aumentar o tamanho efetivo da população, por meio da introdução de animais de outra população, uma vez que o pequeno número de fundadores torna difícil o controle da endogamia.
Palavras chave: Tamanho efetivo da população. Recurso genético. Endogamia. Estrutura de população.
Introduction
There is a deficiency of knowledge about the geographic distributions, representation, economic importance and production of local swine breeds in general, and only a few studies have been performed on the diversity of local swine breeds in South America (McManus et al., 2010). Mariante et al. (2003) reported that the potential of local Brazilian pig breeds has yet to be fully explored, especially with respect to characteristics related to rusticity, adaptability and disease resistance.
The local Brazilian Piau pig is one of the breeds originated from breeds introduced by Portuguese settlers in the 16th century and was influenced by Dutch and African breeds (Vianna, 1985). The breed likely emerged in the southern part of Goiás state, the Triangulo Mineiro region (Minas Gerais state) and western São Paulo state. Piau is the most numerous local breed in the Distrito Federal (Castro et al., 2002). In a census of local Brazilian pig breeds conducted in Paraiba state, only 56 animals were classified as native breeds; among these pigs, sixteen (28.58 %) were classified as Piau, but most of the animals (46.42 %) were classified as belonging to an undefined breed, indicative of a genetic dilution effect (Cavalcante Neto et al., 2007). No census of the Piau breed's occurrence throughout Brazil has been conducted. However, according Mariante et al. (2003) the Piau breed is threatened. In addition, Sollero et al. (2009) found a high rate of inbreeding in two Brazilian herds of Piau pigs using DNA microsatellite analysis.
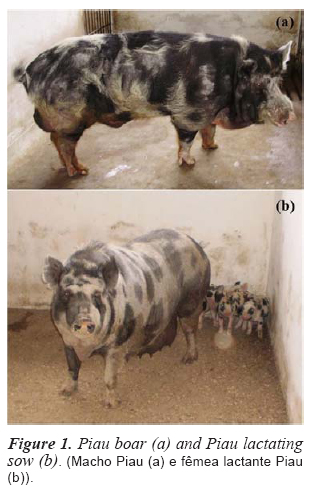
The local Brazilian Piau pig breed is a lard-type pig described by Castro et al. (2000) as having a white or cream-colored coat with black or red spots, smooth and uniformly distributed bristles, a rectilinear or sub-concave head profile and Iberian ear type (figure 1 a and b). The Piau breed underwent a breeding and selection process devised by the veterinarian Antonio Teixeira Vianna in1939. The objective of the program was to develop a dual purpose breed, defined as an animal suitable for the production of both fat and meat (Gomes and D'Aulisio, 1980). The main characteristics of the Piau breed are rusticity, adaptability and disease resistance (Mariante et al., 2003). In recent decades, due to the large amount of fat in their carcasses, their low production efficiency and changes in consumer eating habits, local pig breeds have been threatened with extinction. Some public research institutions and nongovernmental organizations have tried to maintain small herds of these local breeds (Lopes et al., 2002) to preserve their genetic material (Barros et al., 2012a and Barros et al., 2012b) and for genetic studies (Serao et al., 2011; Sollero et al., 2011).
Piau animals have distinct characteristics from those generally favored in intensive production systems. Data from the conservation program of the Federal University of Viçosa reveals that the breed reach sexual maturity between 190 and 210 days, at a body weight of 75 - 80 kg. The sows produce an average of 8.1 ± 2.7 live piglets and 0.9 ± 1.1 stillbirths per litter. The average birth weight is 886.9 ± 130.9 grams. The Piau breed has higher subcutaneous and intramuscular fat deposition than most breeds used in intensive production systems. Serão et al. (2011) reported that Piau pigs weighing 120 kg have a significantly higher intramuscular fat content (2.48 %) compared with a commercial pig formed by a crossing between the breeds Landrace, Large White and Pietrain (1.00 %) at the same weight. The genetics underpinnings of the large fat deposition of Piau pigs have been explored in genetic studies by the Animal Breeding and Genetics group of the Federal University of Viçosa, Minas Gerais state, Brazil. These studies include QTL mapping using divergent crosses (Pires et al., 2005 and 2007; Paixão et al., 2008; Silva et al., 2008), polymorphism analysis of candidate genes (Band et al., 2005; Carmo et al., 2005; Faria et al., 2006; Peixoto et al., 2006), phylogeny and genetic diversity analyses (Sollero et al., 2009; Souza et al., 2009) and expression patterns of candidate genes (Sousa et al., 2011; Serão et al., 2011; Sollero et al., 2011).
Conservation programs are important for maintaining genetic variability, as only a small number of breeds are used for commercial purposes. Retention of genetic diversity is important for overcoming future challenges and meeting consumer needs; moreover, it represents a key part of the cultural and biological heritage of a country or a region (FAO, 2000). Information about population structure is essential for establishing conservation priorities and strategies (Caballero and Toro, 2002). Parameters such as inbreeding, the effective number of founders and effective population size are essential for planning conservation programs and avoiding the loss of genetic variability. An analysis of these parameters provides information about the current status of the breed and aids in planning the next steps of the conservation program.
The objectives of the present study were to describe the Piau genetic conservation program of the Federal University of Viçosa and to evaluate a range of population parameters to generate data for the program's improvement.
Material and methods
In 1998, five males and five females of the local Piau breed were introduced to the Pig Breeding Farm at the Federal University of Viçosa, Minas Gerais state, Brazil, for conservation and genetic research. Over 10 years, populations of approximately 20 females and 5 males were kept for reproduction in each generation. The herd was kept closed until 2008, when two boars from Empresa Baiana de Desenvolvimento Agricola (EBDA) were introduced into the population. Since the introduction of the first animals, mating has been controlled to avoid the loss of genetic variability and inbreeding by mating animals with the lowest coancestry and taking care that all families are represented in the next generation.
Pedigree files containing 1,349 individuals born between 1998 and 2011 were analyzed. Analysis of the population genetic structure was performed with the software ENDOG v 4.8 (Gutierrez and Goyache, 2005), which makes use of the algorithm proposed by Meuwissen and Luo (1992) to compute inbreeding coefficient (F). The inbreeding rate per generation (ΔF) was computed as

Effective population size (Ne) was calculated using the formula:

The average relatedness (AR) was also computed.
The effective number of founders was obtained by

This parameter reflects the number of animals that, given equal contributions, could produce the same degree of genetic variability found in the studied population. The effective number of ancestors a represents the minimum number of animals (founders or not) necessary to explain the total genetic diversity of the evaluated population. This term is complementary to the effective number of founders, as it considers the diversity loss caused by the unbalanced use of individuals, which, in turn, leads to bottlenecks (Gutiérrez and Goyache, 2005). The was a computed by,

Another important parameter for conservation programs is the generation interval, which is defined as the average age of the parents at the birth of their progeny kept for reproduction. This parameter was computed using generation intervals for the four pathways (sire-son, sire-daughter, dam-son and dam-daughter).
The EVA-Inbred software package (Berg, 2003) was used to calculate the expected inbreeding, which is computed as the coancestry of the breeding animals, assuming random mating (Falconer and Mackay, 1996). This measure determines whether appropriate mating strategies have been performed.
Results and discussion
Population parameters are fundamental for the management of genetic diversity and for the planning of a conservation program. Generation intervals for the four pathways (sire-son, sire-daughter, dam-son and dam-daughter) are provided in table I. Generation intervals for all gametic pathways were similar, which means that males and females have been replaced at the same time within the population. Similar generation intervals across gametic pathways are desired in conservation programs because it allows the equal use of both sexes (Barros et al., 2011). Long generation intervals are also desirable because this arrangement results in a lower loss of annual genetic variability and helps to guarantee the higher participation of a given animal (mainly founders) in the population genetic constitution. The generation intervals in the present population (2.557 years) are higher than those found by Welsh et al. (2010), who evaluated five pig breeds in the United States and found generation intervals of 1.65, 1.92, 2.06, 1.83 and 2.21 years for the Berkshire, Duroc, Hampshire, Landrace and Yorkshire breeds, respectively. However, the Piau results are similar to those found by Toro et al. (2000) for Iberian pigs (2.45), which is also a breed kept for conservation. These results show that generation intervals for breeds kept for conservation purposes are divergent from those of selected populations.
In the reference population, there were 1,267 animals for which both parents were known. Twelve ancestors contributed to the formation of this population. Of these animals, four were responsible for 50 % of the population variability, which indicates the unbalanced use of some animals. The effective numbers of founders and ancestors are 9 and 8 animals, respectively. The values of e and a are very small, which was expected because so few animals were used to establish the herd. In addition, the e and a values are similar (9 and 8, respectively), which means that the animals that contributed to the herd formation continue to operate effectively in the current herd. In other words, bottlenecks caused only a small reduction in variability. However, the low values of e and a relative to the number of animals in the reference population implies that the herd evolved from a narrow genetic base, which can result in the loss of original breed genes. Thus, animals from another population should be introduced to increase the genetic base of the population.
Barros et al. (2011), evaluating the Brazilian goat breed Marota, found the effective number of founders and ancestors was 48 animals and the number of ancestors that explained 50 % of population variability was 22. In an analysis of four Canadian pig breeds, Melka and Schenkel (2010) determined the effective number of founders to be 275, 11, 56 and 54 for the Duroc, Hampshire, Lacombe and Landrace breeds, respectively. In addition, the number of ancestors that explained 50 % of the gene pool were especially low for the Hampshire (4 animals) and Lacombe (7 animals) breeds. Thus, problems with small number of founders and populations evolved from narrow genetic base are found in commercial and conservation populations.
Despite the small number of founders, the average inbreeding coefficient of the studied Piau population is not high (6.55 %). Nevertheless, the average relatedness coefficient (AR) is somewhat elevated (19.74 %), which means that the animals from this population are very closely related. This result is expected in a small population, even when minimum coancestry mating is performed; if no action is taken, a further increase in inbreeding in later generations is expected. The individual AR can be used to choose animals for reproduction, such that animals with low AR should be preferred for long-term inbreeding control (Barros et al., 2011).
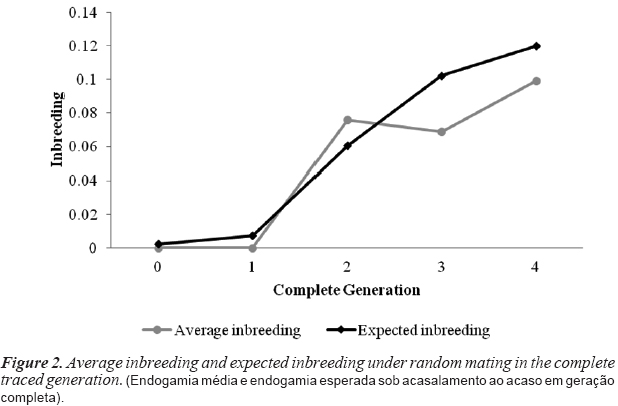
Generation zero is composed of founder animals with unknown pedigrees; thus, the inbreeding of this generation is equal to zero (table II). The increase in average relatedness over subsequent generations can be explained by the population enlargement based on the small initial herd. Inbreeding is equal to zero in the first generation because this population is composed of the founders' progeny. From the second to the third generation, average inbreeding remained almost constant despite the increase in the percentage of inbred animals. This pattern is explained by the considerable growth of the population. In the last (fourth) generation, inbreeding was almost 10 %, and AR was 23 %. This result is worrying because inbreeding is expected to increase further in the next generation. The increase in AR across generations illustrates the loss of genetic diversity, which is troubling because the higher level of AR in the present generation predicts a higher loss of genetic diversity in future generations if no conservation action is taken.
Figure 2 shows the average inbreeding was higher than the expected inbreeding under random mating only in generation two, which reveals that some improper mating was allowed to occur in this generation. In all other generations, the expected inbreeding was higher than the average, showing the effectiveness of the mating strategies for inbreeding control, which were mainly applied in the last two generations. Inbreeding levels and the AR of the current Piau population are shown in figure 3. More than 30 % of the animals have inbreeding coefficients less than 0.025, although, some animals (less than 9 %) have inbreeding coefficients higher than 0.125. These data indicate that while most of the population show low inbreeding, attention should be given to the mating strategy to avoid highly inbred animals in the next generation. A high proportion of the population (77 %) presents average relatedness coefficients between 0.2 and 0.25, indicating that most of the animals are closely related. This result reinforces the conclusion that increased inbreeding and consequent losses of genetic diversity are expected in future generations.
The Food and Agriculture Organization of the United Nations (FAO) (2000) emphasizes that the first objective of a conservation program should be to reach an effective population size (Ne) of 50, which results in an inbreeding rate of 1 % per generation. Nevertheless, Meuwissen and Woolliams (1994) recommended minimum Ne values ranging from 31 to 250 for the maintenance of population fitness. In the studied Piau population, the Ne in generation 4 (table II) is 19.70. This value is below that recommended by the FAO (2000) and Meuwissen and Woolliams (1994). The effective population size of this population should therefore be increased to reduce inbreeding and consequently increase genetic variability. The objective can be accomplished by introducing genetically unrelated individuals. Table II shows the effective population sizes for only generations two and four because it is calculated using the inbreeding rate, which was zero from generation zero to one and negative from generation two to three. table III shows the effective population sizes and inbreeding rates for the fully traced, maximum number and equivalent complete generations, which represent the lower, upper and true limits of Ne, respectively (table III). The upper and lower limits of Ne for the pedigreed population were 68.97 and 18.59, respectively. Despite the upper limit being above 50, the lower limit is well below the recommended threshold value, supporting the recommendation to increase the effective population size. In an analysis of four Canadian pig breeds Melka and Schenkel (2010) found small effective population sizes for the Hampshire (14) and Lacombe (22) breeds and emphasized that these breeds should be the subjects of conservation practices, as a small effective population size will lead to lower genetic diversity in the future. These findings highlight the importance of larger effective population sizes for the maintenance of genetic variability within a breed.
Conclusion
The analyzed population parameters reveal that the conservation program has maintained a long generation interval, which is desirable for conservation purposes. In addition, the average generation intervals are similar for males and females which allows the equal use of both sexes. The effective population size should be increased to preserve genetic variability. This can be accomplished by introducing genetically unrelated individuals, which will also serve to increase the genetic base of the population.
References
1. Band, G.O; Guimarães, S.E.F; Lopes, P.S.; Schierholt, A.S.; Silva, K.M.; Pires, A.V.; Benevenuto Jr., A.A. and Gomide, L.A.M. 2005. Relationship between the porcine stress syndrome gene and pork quality trait in F2 pigs resulting from divergent crosses. Genet Mol Biol, 28: 88-91. [ Links ]
2. Barros, E.A.; Ribeiro, M.N.; Almeida, M.J.O. e Araújo, A.M. 2011. Estrutura populacional e variabilidade genética da raça caprina Marota. Arch Zootec, 60: 543-552. [ Links ]
3. Barros, M.H.C.; Shiomi, H.H.; Amorim, L.S.; Guimarães, S.E.F.; Lopes, P.S.; Siqueira, J.B.; Pinho, R.O.; Pereira, J.V.T.N. e Guimarães, J.D. 2012a. Características quantitativas e qualitativas do sêmen in natura de suínos da raça Piau. Rev Cient Eletrôn Med Vet, http://www.revista.inf.br/veterinaria18/artigos/art13.pdf (28/05/2012). [ Links ]
4. Barros, M.H.C.; Amorim, L.S.; Guimarães, S.E.F.; Lopes, O.S.; Siqueira, J.B. e Guimarães, J.D. 2012b. Criopreservação de sêmen de suínos da raça Piau (Sus scrofa) submetido a três protocolos de congelamento. Rev Bras Zootecn, 41: 914-922. [ Links ]
5. Berg, P. 2003. EVA version 1.4. Evolutionary algorithm for mate selection. User's guide. Danish Institute of Agricultural Sciences. Foulum. Denmark. [ Links ]
6. Caballero, A. and Toro, M.A. 2002. Analysis of genetic diversity for the management of conserved subdivided populations. Conserv Genet, 3: 289-299. [ Links ]
7. Carmo, F.M.; Guimarães, S.E.F.; Lopes, P.S.; Pires, A.V.; Guimarães, M.F.M.; Silva, M.V.G.B.; Schierholt, A.S.; Silva, K.M. and Gomide, L.A.M. 2005. Association of MYF5 gene allelic variants with production traits in pigs. Genet Mol Biol, 28: 363-369. [ Links ]
8. Castro, S.T.R.; Albuquerque, M.S.M. and Germano, J.L. 2000. Conservation of local pig breeds in Brasil. Proc. Fifty Global Conference on Conservation of Domestic Animal Genetic Resources. Embrapa Recursos Genéticos e Biotecnologia. Brasília, DF. Brasil. [ Links ]
9. Castro, S.T.R.; Albuquerque, M.S.M. and Germano, J.L. 2002. Census of Brazilian naturalized swine breeds. Arch Zootec, 51: 235-239. [ Links ]
10. Cavalcante Neto, A.; Silva, L.P.G.; Ribeiro, M.N.; Lui, J.F.; Barbosa, J.G.; Castro, S.T.R. and Souza, G.J.G. 2007. Censo e caracterização fenotípica de suínos de grupos genéticos naturalizados brasileiros existentes no Estado da Paraíba. Biotemas, 20: 123-126. [ Links ]
11. Falconer, D.S. and Mackay, T.F.C. 1996. Introduction to quantitative genetics. 4th ed. Longman Scientific and Technical. Harlow, UK. [ Links ]
12. FAO. 2000. Secondary guidelines for development of national farm. Animal genetic resources management plans: Management of small populations at risk. Organização das Nações Unidas para Agricultura e Alimentação. Rome, Italy. [ Links ]
13. Faria, D.A.; Guimarães, S.E.F.; Lopes, P.S.; Pires, A.V.; Paiva, S.R.; Sollero, B.P. and Wenceslau, A.A. 2006. Association between G316A growth hormone polymorphism and economic traits in pigs. Genet Mol Biol, 29: 634-640. [ Links ]
14. Gomes, M.B. e D'Aulísio, S.H.G. 1980. Estudo da prolificidade da raça suína Piau. An Esc Super Agric Luiz de Queiroz, 37: 179-208. [ Links ]
15. Gutiérrez, J.P. and Goyache, F. 2005. A note on ENDOG: a computer program for analyzing pedigree information. J Anim Breed Genet, 122: 172-176. [ Links ]
16. Lopes, P.S.; Guimarães, S.E.F.; Pires, A.V.; Soares, M.A.M.; Carmo, F.M.S.; Martins, M.F.; Benevenuto Júnior, A.A. and Gomide, L.A.M. 2002. Results of performance, carcass yield and meat quality traits of F2 crosses between Brazilian native and commercial pigs for QTL mapping. Proc. 7th World Congress on Genetics Applied to Livestock Production. Montpellier, France. [ Links ]
17. Mariante, A.S.; Castro, S.T.R.; Albuquerque, M.S.M.; Paiva, S.R. and Gemano, J.L. 2003. Pig biodiversity in Brazil. Arch Zootec, 52: 245-248. [ Links ]
18. Melka, M.G. and Schenkel, F. 2010. Analysis of genetic diversity in four Canadian swine breeds using pedigree data. Can J Anim Sci, 90: 331-340. [ Links ]
19. McManus, C.; Paiva, S.R.; Silva, A.V.R.; Murata, L.S.; Louvandini, H.; Cubillos, G.P.B.; Castro, G.; Martinez, R.A.; Dellacasa, M.S.L. and Perez, J.E. 2010. Phenotypic characterization of naturalized swine breeds in Brazil, Uruguay and Colombia. Braz Arch Biol Technol, 53: 583-591. [ Links ]
20. Meuwissen, T.H.E and Luo, Z. 1992. Computing inbreeding coefficients in large populations. Genet Sel Evol, 24: 305-313. [ Links ]
21. Meuwissen, T.H.E. and Woolliams, J.A. 1994. Effective sizes of livestock populations to prevent a decline in fitness. Theor Appl Genet, 89: 1019-1026. [ Links ]
22. Paixão, D.M.; Silva Filho, M.I.; Pereira, M.S.; Lopes, M.S.; Barbosa, L.; Souza, K.R.S.; Lopes, P.S. and Guimarães, S.E.F. 2008. Detection of quantitative trait loci on chromosomes 16, 17 and 18 for carcass, internal organs and meat quality traits is pigs. Genet Mol Biol, 31: 98-901. [ Links ]
23. Peixoto, J.O.; Guimarães, S.E.F.; Lopes, P.S.; Soares, M.A.M.; Pires, A.V.; Barbosa, M.V.G.; Torres, R.A. and Silva, M.A. 2006. Associations of leptin gene polymorphisms with production traits in pigs. J Anim Breed Genet, 123: 378-383. [ Links ]
24. Pires, A.V.; Lopes, P.S.; Guimarães, S.E.F; Guimarães, C.T.; Gomide, L.A.M.; Benevenuto Jr., A.A. and Carmo, F.M.S. 2005. Quantitative trait loci mapping for meat quality traits in swine chromosome 6. Arq Bras Med Vet Zootec, 57: 608-615. [ Links ]
25. Pires, A.V.; Lopes, P.S.; Guimarães, C.T. and Guimarães, S.E.F. 2007. Mapping quantitative trait loci for performance traits on pig chromosome 6 (SSC6). Arch Latinoam Prod Anim, 15: 25-32. [ Links ]
26. Serão, N.V.L.; Veroneze, R.; Ribeiro, A.M.F.; Verardo, L.L.; Braccini Neto, J.; Gasparino, E.; Campos, C.F.; Lopes, P.S. and Guimarães, S.E.F. 2011. Candidate gene expression and intramuscular fat content in pigs. J Anim Breed Genet, 128: 29-34. [ Links ]
27. Silva, K.M.; Paixão, D.M.; Silva, P.V.; Sollero, B.P.; Pereira, M.S.; Lopes, P.S. and Guimarães, S.E.F. 2008. Mapping of quantitative trait loci and confirmation of the FAT1 region on chromosome 4 in an F2 population of pigs. Genet Mol Biol, 31: 475-480. [ Links ]
28. Sollero, B.P.; Paiva, S.R.; Faria, D.A.; Guimarães, S.E.F.; Castro, S.T.R.; Egito, A.A.; Albuquerque, M.S.M.; Piovesan, U.; Bertani, G.R. and Mariante, A.S. 2009. Genetic diversity of Brazilian pig breeds evidenced by microsatellite markers. Livest Sci, 123: 8-15. [ Links ]
29. Sollero, B.P.; Guimarães, S.E.F.; Rilington, V.D.; Tempelman, R.J.; Raney, N.E.; Steibel, J.P.; Guimarães, J.D.; Lopes, P.S.; Lopes, M.S. and Ernst, C.W. 2011. Transcriptional profiling during fetal skeletal muscle development of Piau and Yorkshire-Landrace cross-bred pigs. Anim Genet, 42: 600-612. [ Links ]
30. Sousa, K.R.S.; Ribeiro, A.M.F.; Goes, P.R.N.; Guimarães, S.E.F.; Lopes, P.S.; Veroneze, R. and Gasparino, E. 2011. Toll-like receptor 6 differential expression in two pig genetic groups vaccinated against Mycoplasma hyopneumoniae. BMC Proc, 5 (Suppl 4): S9. [ Links ]
31. Souza, C.A.; Paiva, S.R.; Pereira, R.W.; Guimarães, S.E.F.; Dutra Jr., W.M.; Murata, L.S. and Mariante, A.S. 2009. Iberian origin of Brazilian local pig breeds based on Cytochrome b (MT-CYB) sequence. Anim Genet, 40: 759-762. [ Links ]
32. Toro, M.A.; Rodrigañez, J.; Silio, L. and Rodriguez, C. 2000. Genealogical analysis of a closed herd of Black Hairless Iberian pigs. Conserv Biol, 14: 1843-1851. [ Links ]
33. Vianna, A.T. 1985. Os suínos. Editora Nobel. São Paulo, Brazil. [ Links ]
34. Welsh, C.S.; Stewart, T.S.; Schwab, C. and Blackburn, H.D. 2010. Pedigree analysis of 5 swine breeds in the United States and the implications for genetic conservation. J Anim Sci, 88: 1610-1618. [ Links ]
Recibido: 21-9-12
Aceptado: 25-9-13












 Curriculum ScienTI
Curriculum ScienTI