Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO  Similares en Google
Similares en Google
Compartir
Revista Española de Enfermedades Digestivas
versión impresa ISSN 1130-0108
Rev. esp. enferm. dig. vol.97 no.3 Madrid mar. 2005
| ORIGINAL PAPERS |
Overexpression of c-myc and loss of heterozigosity on 2p, 3p, 5q, 17p and 18q in sporadic colorectal carcinoma
A. Sánchez-Pernaute, E. Pérez-Aguirre, F.J. Cerdán, P. Iniesta1, L. Díez Valladares, C. de Juan1, A. Morán1, A. García-Botella,
C. García Aranda1, M. Benito1, A.J. Torres and J.L. Balibrea
Department of Surgery. Hospital Universitario San Carlos. Universidad Complutense de Madrid. 1Department of Molecular Biology
and Biochemistry. Pharmacy Faculty. Universidad Complutense de Madrid. Spain
ABSTRACT
Aim: the aim of the present study is to evaluate the prognostic influence of loss of heterozygosity on 2p, 3p, 5q, 17p and 18q, and c-myc overexpression on surgically treated sporadic colorectal carcinoma.
Methods: tumor and non-tumor tissue samples from 153 patients were analyzed. Fifty-one percent of patients were male, and mean age in the series was 67 years. Tumors were located in the proximal colon in 37 cases, in the distal bowel in 37, and in the rectum in 79 patients. c-myc overexpression was studied by means of Northern blot analysis, and loss of heterozigosity through microsatellite analysis.
Results: c-myc overexpression was detected in 25% of cases, and loss of heterozygosity in at least one of the studied regions in 48%. There was no association between clinical and pathologic features, and genetic alterations. The disease-free interval was significantly shorter for patients with both genetic alterations; the presence of both events was an independent prognostic factor for poor outcome in the multivariate analysis (RR: 4.34, p < 0.0001).
Conclusions: the presence of both loss of heterozygosity and overexpression of the c-myc oncogene separates a subset of colorectal carcinoma patients who have a shorter disease-free interval after curative-intent surgery.
Key words: Loss of heterozygosity. c-myc. Oncogenes. Prognosis. Colorectal carcinoma.
This study has been partially financed by the project FIS 94/1557 and the project "Genetic Instability in Colorectal Carcinoma", Aventis-Pharma S.A.
Recibido: 14-06-04.
Aceptado: 05-10-04.
Correspondencia: Andrés Sánchez-Pernaute. Departamento de Cirugía, 7ª planta, ala Norte. Hospital Clínico San Carlos. C/ Martín Lagos, s/n. 28040 Madrid. e-mail: andresanchez@hotmail.com
INTRODUCTION
Colorectal carcinoma (CRC) is one of the most frequently diagnosed solid tumors. It is the second cause of cancer-related death in the Western world (1,2), and in Spain it causes 11% of cancer-related deaths in the male population and 15% in the female one (3).
The high incidence of CRC and the ease with which tumor tissue samples are obtained in different evolutionary stages have both led to the description of a tumor progression model, from benign polyps to invasive cancers (4), all along the genetic events associated with this neoplastic transformation. These include, APC deletion, DNA hypomethylation, k-ras mutations, allelic losses in DCC, or p53 alterations (4,5).
The expression of some of these genetic alterations seems to accompany tumoral phenotypes -more or less aggressive- and the detection of these alterations could be in the future the basis for deciding the application of adjuvant therapies, which to date are only based in the only universally accepted prognostic factor -tumor stage.
The present work analyzes the overexpression of the c-myc gene, a dominant oncogene that may also behave as a suppressor gene, and the loss of heterozygosity in different chromosomal regions in a series of surgically-treated patients with sporadic CRC, as well as the influence of these alterations on the recurrence of disease.
PATIENTS AND METHODS
Patients
One-hundred and fifty-three patients, 78 males and 75 females, electively operated on for sporadic CRC, were included. Mean age in this series was 67 years (31-89). Eighteen patients had a Dukes' A tumor, 61 had a Dukes' B, 47 a Dukes' C, and 27 had a stage D. Thirty-seven tumors were located in the proximal colon, 37 in the distal bowel, and 79 in the rectum.
Three patients (2%) died in the postoperative period and were excluded of the survival analysis.
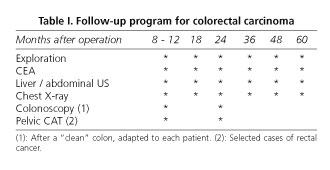
Patients with stage-B or higher disease were postoperatively treated with adjuvant therapy unless medically contraindicated. After discharge or chemotherapy completion, all patients were included in a follow-up protocol (6) (Table I). Suspicion of tumor relapse guided the intent to obtain cytological or histological confirmation, but a radiolographic image with or without elevated tumor markers was enough to diagnose tumor recurrence.
Sample harvesting. Immediately after bowel resection, the specimen was opened and washed. Tumor and normal mucosa samples were harvested. Tissue specimens were frozen in liquid nitrogen and stored at -80 ºC in a freezer for later analysis. All patients gave their informed consent for the use of their tissues, and the study was approved by the local Ethics Committee.
c-myc overexpression. c-myc overexpression was evaluated using a Northern-blot analysis. Tissues for RNA extraction, frozen at -80 ºC, were immediately homogenized in an ice-cold solution of guanidinium thyocyanate, to minimize the risk of RNA degradation. Moreover, all solutions used for RNA extraction were RNAase-free. Thus, RNA was extracted from tumor and non-tumor samples using Chomczynski and Sacchi method (7), and quantitated by UV spectrophotometry. Thirty micrograms of total RNA were denatured in 50% formamide, 2.2 M formaldehyde, 20 mM MOPS (morpholin propanosulphonic acid) (pH 7.0), 6% glycerol at 65 ºC for 15 min, separated by size on gels containing 0.9% agarose and 0.66 M formaldehyde, and blotted on GeneScreenTM membranes (NEN Research Products, Dupont, Boston, MA). RNA was cross-linked to the membrane by UV light. The membrane was then prehybridized for 3 hours and hybridized for 24 h at 42 ºC in a buffer containing 0.25 M Na2PO4 (pH 7.2), 0.25 M NaCl, 1 mM EDTA, 100 mg/ml denatured salmon sperm DNA, 7% SDS (sodium dodecyl sulphate), 50% deionized formamide, and the labeled probe. The c-myc DNA probe was labeled with (α32P) dCTP by random priming reaction. After hybridization, membranes were washed and exposed to Kodak X-OMAT AR films. Densitometric quantification of the autoradiograms was performed in a Molecular Dynamics (Sunnyvale, CA) scanning laser densitometer. The 18S ribosomal probe was used for RNA normalization. Results were obtained comparing the c-myc signal in tumor samples to the c-myc signal in non-tumor samples.
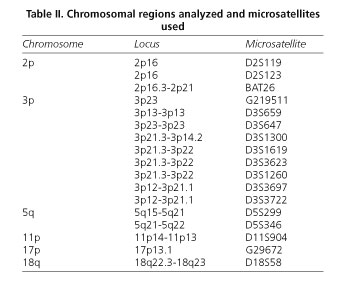
Loss of heterozigosity. DNA was extracted as reported by Blin and Stafford (8). DNA samples were analyzed by polymerase chain reaction for loss of heterozigosity at loci exposed in table II. The polymerase chain reaction was performed in a Thermocycler Perkin Elmer (Gene Amp PCR System 2400) system, and was carried out in a 20 μl volume containing: 50 ng of genomic DNA, 1 μM of primer, 0.2 mM of each deoxynucleoside triphosphate, 10 mM Tris HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 0.5 ml (α32P)dCTP (3000 Ci/mmol) (Amersham, Buckinghamshire, UK), and 1 U of Dynazyme Thermostable DNA Polymerase (Finnzymes OY, Helsinki, Finland). PCR conditions were as follows: initial denaturation at 94 ºC for 5 min, 30 cycles of 94 ºC for 0.5 min, 55 ºC for 0.5 min, and 72 ºC for 1 min. Final extension was 72 ºC for 7 min. PCR products were denatured by 95% formamide and electrophoresed on 7 M urea polyacrylamide gels for 3 h at 43 W, followed by autoradiography. Loss of heterozygosity was considered when a complete loss of one or both alleles of the locus appeared.
Statistical analysis. All clinical and pathological results were compared with molecular data by means of χ2 or Fisher's tests for qualitative variables, and t-test for continuous ones. Disease-free survival was studied with Kaplan Meier curves, compared with the log rank test. Variables obtaining prognostic signification were entered into a Cox Proportional Hazards Model. All calculations were carried out with the Statistical Package for Social Sciences, version 10.0, for Windows (SPSS Inc, Chicago, IL, license number 7269515).
RESULTS
In 39 cases, we detected overexpression of c-myc (25.5%), with no relationship to sex, tumor location, grade of differentiation, or tumor stage. Patients with c-myc overexpression were significantly younger, 64.2 vs. 68.6 years, p = 0.03.
We could study loss of heterozigosity (loh) in 143 samples; in 69 cases (48.3%) loh was detected in at least one of the studied regions. There was no association to any of the variables analyzed, but we found a non-statistical association between loh and grade of differentiation, as 32% of well-differentiated tumors showed loh, 51% of intermediate ones harboured the alteration, and 57% of poorly differentiated tumors were loh+ (p = 0.09).
Among 143 patients in which both genetic alterations were analyzed, 57 cases (39.9%) were negative both for loh and c-myc overexpression, and 20 cases were positive for both of them.
Survival studies
Mean follow-up was 28 months, with a range from 0 to 115 months. We detected 35 tumor recurrences (22.9%). Mean disease-free survival for patients in stages A to C was 69% at 5 years. c-myc overexpression was associated with a shorter disease-free survival, mainly between patients in Dukes' B stage (c-myc +: 58% vs. c-myc -: 86%), but statistical signification was not reached.
Loh was a poor prognosis feature, as the disease-free survival estimation was 56% at 5 years for patients with allelic loss, while it was 75% for patients without loh (p = 0.07); differences were higher in Dukes' B stage (68 vs. 92%, p < 0.05), reaching statistical signification.
In an attempt to determine the prognostic influence of both alterations together, the series was divided into four subsets: patients with no detectable alterations, patients with c-myc overexpression and no loh, patients harbouring loh but not c-myc overexpression, and patients with both genetic alterations. Disease-free survival showed a significantly worse prognosis for the latter group -patients with both c-myc overexpression and loh- as only 33% of these patients were disease-free at 5 years (p = 0.002) (Fig. 1).
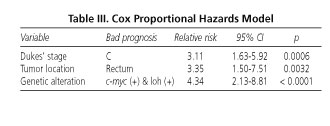
To estimate prognostic independence, genetic alterations were compared to "classic" prognostic factors such as Dukes' stage and tumor location. In the multivariate analysis all three behaved as independent prognostic factors, with the relative risk for recurrence being 3.11 for patients in stage C, 3.35 for tumors located in the rectum, and 4.34 for tumors with loh and c-myc overexpression (Table III).
DISCUSSION
The c-myc oncogene is overexpressed or amplified in one third of CRCs (9,10), and in up to 65% of cases when its protein is investigated by means of immunohistochemical analysis (11). The difference between DNA or RNA analysis and protein analysis lies in that the former technique only evaluates increased synthesis, while the latter includes also decreased degradation. It is not clear whether the accumulation of c-myc is a feature of more advanced tumors as communicated by Masramon et al. (9) and Heerdt et al. (12), or if, as we have presented, the overexpression of this gene develops in the initial stages of tumor progression, being independent of tumor stage (11,13). Attending to the location of the tumor, most authors find increased expression of c-myc in distal colon tumors and rectal cancers, while proximal tumors would be characterized by other genetic alterations such as microsatellite instability (14,15).
Loh has been reported in up to 80% of CRCs. Loh refers to the alteration detected as a partial or total loss of one of the alleles of the analyzed loci, and we define the genotype loh +, which appears to characterize a different phenotype of colorectal cancer (16). In our study we investigated all the chromosomal regions involved in CRC, i.e. 2p, 3p, 5q, 17p and 18q, which are those containing already identified -or unidentified but suspected as yet (2p, 3p)- suppressor genes (APC, p53, DCC). Loh is reported more frequently in advanced tumors, as if there was a progressive loss of genetic material parallel to tumor progression (17); in our series, loh prevalence is just below 50%, and it appears not to be associated with tumor stage or the location of the tumor (18).
The prognostic value of c-myc overexpression in CRC has been studied in many different ways, but neither DNA amplification (19), nor RNA overexpression (20,21) or the immunohistochemical analysis of the protein (11,13) have yet clarified the role of this oncogene in the evolution of this disease. Augenlicht et al. (19) reported a very interesting work where they found that patients with c-myc gene amplification had a better response to surgery plus adjuvant therapy with levamisole and 5-fluorouracil. Although their series is short and they did not find prognostic independence, their paper represents a good basis for the investigation of response to chemotherapy in association with the expression of this gene. Bhatavdekar et al. (11) found prognostic differences in relation to c-myc protein expression in a short series of Dukes' B and C patients, with a greater level of significance when c-myc was considered together with Bcl-2 and p53; the authors postulate that the prognostic importance of these genes lies in the cooperation between them. The same conclusions are reported by Smith and Goh (21), who found a different prognosis -in this case a better one- in the group of patients with c-myc overexpression associated with "wild-type" p53 overexpression.
Chromosomal deletions usually affect negatively the prognosis of CRC. Our group recently reported a relationship between 3p23 loh and shorter survival (22). In the case of 17p the prognostic influence is variable (23), and one can even find reports that confer a protective value to p53 alterations (24,25). Allelic losses in 18q are usually found to be related to a poorer clinical course, as reported by Vogelstein et al. (26) and by Rosell et al. in our country (27); both studies find 18q loh to be an independent prognostic factor for CRC. The same is found for 5q loh (APC), which usually behaves as a negative prognostic attribute (28).
Taken all allelic losses together, i.e. considering "loh +" the phenotype characterized by the presence of at least one allelic loss, we find a worse prognosis for patients harbouring this anomaly, just as our group and other authors have previously reported (17,29). Though statistical significance is not reached, when stage B is considered separately differences reach signification, which almost happens in the case of c-myc. This is a quite important fact, as stage B patients are those who obtain no clear benefit from adjuvant therapy; with a genetic study, we could select patients with an estimated poor outcome, and they would receive postoperative chemotherapy.
The association of both genetic events is clearly a poor prognostic factor, and it behaves independently regarding other known prognostic variables. How could both genetic events be linked? The oncogene c-myc was originally described as a gene activated following mutagenic stimuli, depending upon the expression of growth factors and with a regulatory activity on cell growth. Its presence seems to be essential for the progression from phase G1 to S, and it also regulates cellular differentiation and promotes transformation.
It has recently been rediscovered, as a tumor suppressor gene that may induce cell death through apoptosis. The dual function depends upon the cellular environment and the status of the genes targeted by c-myc (30). Regarding cooperation mechanisms, or the relationship between c-myc and the deleted genes in the loh genotype, some of them have already been elucidated. The APC gene in 5q is connected to c-myc through the wnt way, a membrane protein implied in intercellular communication, growth and differentiation. Wnt signals induce the release of the complex APC-b-catenin, and so b-catenin can move to the nucleus to inhibit the suppression of c-myc by different proteins such as Groucho or CBP. In this way stimulus from the wnt receptor induces cellular proliferation and transformation through c-myc, and this mechanism is reproduced when there is APC mutation or 5q deletion, as b-catenin cannot complete its action (31-33). On the other hand, cooperation with p53 appears to occur through the opposite way, the negative growth control or its behaviour as an apoptotic inductor and a tumor suppressor gene. It seems that "wild-type" p53 protein plays a role in Fas-mediated apoptosis as induced by c-myc (34). In this way, gene mutations or 17p deletions would cause an inhibition of c-myc -mediated apoptosis, a stimulation of cellular transformation, and neoplasic development. There are probably similar mechanisms which positively or negatively relate to the different genes considered in the loh genotype, and they would be responsible for the poorer outcome of patients harbouring these alterations.
REFERENCES
1. Greenle RT, Murray T, Bolden S, Wingo PA. Cancer statistics, 2000. CA Cancer J Clin 2000; 50: 7-33. [ Links ]
2. Cohen AM, Minsky BD, Schilsky RL. Cancer of the colon. In: DeVita Jr VT, Hellman S, Rosenberg SA. Cancer. Principles and practice of oncology. 5th ed. Philadelphia: Lippincott - Raven Publishers, 1997. p. 1144-97. [ Links ]
3. Centro Nacional de Epidemiología. Área de Epidemiología Ambiental y Cáncer. Mortalidad por Cáncer en España, 2000. http://cne.isciii.es/cancer/mort2000.txt. [ Links ]
4. Fearon ER, Vogelstein B. A genetic model for colorectal carcinogenesis. Cell 1990; 61: 759-67. [ Links ]
5. Toribara NW, Slisenger MH. Screening for colorectal cancer. N Engl J Med 1995; 332: 861-7. [ Links ]
6. Cerdán FJ. Seguimiento de los pacientes intervenidos de cáncer colorrectal. Rev Cancer 1997; 11: 32-41. [ Links ]
7. Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987; 162: 156-9. [ Links ]
8. Blin N, Stafford DW. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acid Res 1976; 3: 2303-8. [ Links ]
9. Masramon L, Arribas R, Tórtola S, Perucho M, Peinado MA. Moderate amplifications of the c-myc gene correlate with molecular and clinicopathological parameters in colorectal cancer. Br J Cancer 1998; 99: 2349-56. [ Links ]
10. Arango D, Corner GA, Wadler S, Catalano PJ, Augenlicht LH. C-myc/p53 interaction determines sensitivity of human colon carcinoma cells to 5-fluorouracil in vitro and in vivo. Cancer Res 2001; 61: 4910-5. [ Links ]
11. Bhatavdekar JM, Patel DD, Ghosh N, Chikhlikar PR, Trivedi TI, Suthar TP, et al. Coexpression of Bcl-2, c-Myc and p53 oncoproteins as prognostic discriminants in patients with colorectal carcinoma. Dis Colon Rectum 1997; 40: 785-90. [ Links ]
12. Heerdt BG, Molinas S, Deitch D, Augenlicht LH. Aggressive subtypes of human colorectal tumors frequently exhibit amplification of the c-myc gene. Oncogene 1991; 6: 125-9. [ Links ]
13. Miller F, Heimann TM, Quish A, Pyo DJ, Szporn A, Martinelli G, et al. Ras and c-myc protein expression in colorectal carcinoma. Study of cancer prone patients. Dis Colon Rectum 1992; 35: 430-5. [ Links ]
14. Rothberg G, Spandorfer JM, Erisman MD, Staroscik RN, Sears HF, Petersen RO, et al. Evidence that c-myc expression defines two genetically distinct forms of colorectal adenocarcinoma. Br J Cancer 1985; 52: 629-32. [ Links ]
15. Bufill JA. Colorectal cancer: Evidence for distinct genetic categories based on proximal or distant tumor location. Ann lntern Med 1990; 113: 779-88. [ Links ]
16. Goel A, Arnold CN, Niedzwiecki D, Chang DK, Ricciardiello L, Carethers JM, et al. Characterization of sporadic colon cancer by patterns of genomic instability. Cancer Res 2003; 63: 1608-14. [ Links ]
17. Weber JC, Schneider A, Rohr S, Nakano H, Bachellier P, Méchine A, et al. Analysis of allelic imbalance in patients with colorectal cancer according to stage and presence of synchronous liver metastases. Ann Surg 2001; 234: 795-803. [ Links ]
18. García-Hisrschfeld García J, Blanes Berenguel A, Vicioso Recio L, Márquez Moreno A, Rubio Garrido J, Matilla Vicente A. Colon cancer: p53 expression and DNA ploidy. Their relation to proximal or distal tumor site. Rev Esp Enferm Dig 1999; 91: 481-8. [ Links ]
19. Augenlicht LH, Wadler S, Corner G, Richards C, Ryan L, Multani AS, et al. Low-level c-myc amplifications in human colonic carcinoma cell lines and tumours: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res 1997; 57: 1769-75. [ Links ]
20. Nagai MA, Habr-Gama A, Oshima CT, Brentani MM. Association of genetic alterations of c-myc , c-fos, and c-Ha-ras proto-oncogenes in colorectal tumors. Frequency and clinical significance. Dis Colon Rectum 1992; 35: 444-51. [ Links ]
21. Smith DR, Goh HS. Overexpression of the c-myc proto-oncogene in colorectal carcinoma is associated with a reduced mortality that is abrogated by point mutation of the p53 tumor suppressor gene. Clin Cancer Res 1996; 2: 1049-53. [ Links ]
22. lniesta P, Massa MJ, González-Quevedo R, de Juan C, Morán A, Sánchez-Pernaute A, et al. Loss of heterozygosity at 3p23 is correlated with poor survival in patients with colorectal carcinoma. Cancer 2000; 89: 1220-7. [ Links ]
23. Petersen S, Thames HD, Nieder C, Petersen C, Baumann M. The results of colorectal cancer treatment by p53 status: treatment-specific overview. Dis Colon Rectum 2001; 44: 322-34. [ Links ]
24. Soong R, Grieu F, Robbins P, Dix B, Chen D, Parsons R, et al. p53 alterations are associated with improved prognosis in distal colonic carcinomas. Clin Cancer Res 1997; 3: 1405-11. [ Links ]
25. Ahnen DJ, Feigi P, Quan G, Fenoglio-Preiser C, Lovato LC, Bunn Jr PA, et al. Ki-ras mutations and p53 overexpression predict the clinical behavior of colorectal cancer: a Southwest Oncology Group Study. Cancer Res 1998; 58: 1149-58. [ Links ]
26. Jen J, Kim H, Piantadosi S, Liu Z-F, Levitt RC, Sistonen P, et al. Allelic loss of chromosome 18q and prognosis in colorectal cancer. N Engl J Med 1994; 331: 213-21. [ Links ]
27. Martínez-López E, Abad A, Font A, Monzó M, Ojanguren I, Pifarré A, et al. Allelic loss on chromosome 18q as a prognostic marker in stage II colorectal cancer. Gastroenterology 1998; 114: 1180-7. [ Links ]
28. McLeod HL, Murray GI. Tumour markers of prognosis in colorectal cancer. Br J Cancer 1999; 79: 191-203. [ Links ]
29. Massa MJ, lniesta P, González Quevedo R, de Juan C, Caldés T, Sánchez- Pernaute A, et al. Differential prognosis of replication error phenotype and loss of heterozigosity in sporadic colorectal cancer. Eur J Cancer 1999; 35: 1676-82. [ Links ]
30. Packham G, Cleveland JL. c-Myc and apoptosis. Biochem Biophys Acta 1995; 1242: 11-28. [ Links ]
31. Pennisi E. How a growth control path takes a wrong turn to cancer. Science 1998; 281: 1438-41. [ Links ]
32. He T-C, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, et al. Identification of C-MYC as a target of the APC pathway. Science 1998; 281: 1509-12. [ Links ]
33. Seidler HBK, Utsuyama M, Nagaoka S, Takemura T, Kitagawa M, Hirokawa K. Expression level of Wnt signaling components possibly influences the biological behavior of colorectal cancer in different age groups. Exp Mol Pathol 2004; 76: 224-33. [ Links ]
34. Green DR. A Myc-induced apoptosis pathway surfaces. Science 1997; 278: 1246-7. [ Links ]











 texto en
texto en