My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Española de Enfermedades Digestivas
Print version ISSN 1130-0108
Rev. esp. enferm. dig. vol.107 n.11 Madrid Nov. 2015
Role of colonic microbiota in colorectal carcinogenesis: A systematic review
Marta Borges-Canha1, José Pedro Portela-Cidade1, Mário Dinis-Ribeiro2,3, Adelino F. Leite-Moreira1 and Pedro Pimentel-Nunes1,2,3
1 Department of Physiology and Cardiothoracic Surgery, Cardiovascular Research & Development Unit. Faculty of Medicine. University of Porto. Portugal.
2 Gastroenterology Department. Portuguese Oncology Institute. Porto, Portugal.
3 CINTESIS/Department of Biostatistics and Medical Informatics. Porto Faculty of Medicine. Porto, Portugal
ABSTRACT
Background and aim: The human colonic mucosa is populated by a wide range of microorganisms, usually in a symbiotic relation with the host. Sometimes this balance is lost and a state of dysbiosis arises, exposing the colon to different metabolic and inflammatory stimuli (according to the microbiota's changing profile). Recent findings lead to hypothesize that this unbalance may create a subclinical pro-inflammatory state that increases DNA mutations and, therefore, colorectal carcinogenesis. In this article we aim to systematically review the scientific evidence regarding colonic microbiota and its role in colorectal carcinogenesis.
Methods: Systematic review of PubMed searching results for original articles studying microbiota and colorectal cancer until November 2014.
Results: Thirty-one original articles studied the role of colon microbiota in colorectal carcinoma including both human and animal studies. Different and heterogeneous methods were used and different bacteria were considered. Nevertheless, some bacteria are consistently augmented (such as Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteridae, Staphylococcaceae, Akkermansia spp. and Methanobacteriales), while other are constantly diminished in colorectal cancer (such as Bifidobacterium, Lactobacillus, Ruminococcus, Faecalibacterium spp., Roseburia, and Treponema). Moreover, bacteria metabolites amino acids are increased and butyrate is decreased throughout colonic carcinogenesis.
Conclusion: Conclusive evidence shows that colorectal carcinogenesis is associated with microbial dysbiosis. This information may be used to create new prophylactic, diagnostic and therapeutic strategies for colorectal cancer.
Key words: Colon microbiota. Microbiome. Colorectal cancer. Innate immunity.
Introduction
The human large bowel is known for its wide microbiota composition. In fact, there are as many as 100 trillion organisms that interfere with the host, usually in symbiotic relation. These microorganisms take the undigested nutrients that reach the colon as its substrates to live. Usually, those are innocuous commensals or are relevant to final product degradation, as well as vitamin formation, among other functions (1-3).
However, this balance is not always maintained and the chronic inflammation and immune evasion caused by inappropriate interactions may promote colorectal carcinogenesis. Indeed, there is growing evidence on microbial dysbiosis in colorectal cancer patients, although the mechanism is not fully understood and yet to be investigated (4,5).
Colorectal cancer (CRC) is a troublesome issue because it is a major cause of cancer deaths around the world, mostly in developed countries where its incidence is increasing. It is a multifactorial disease, associated with lifestyle (pointing out dietary habits and sedentary behaviuors), DNA mutations, inflammation and, most recently, microbiota changes (6-8).
Although advances are still minimal, recent researches have attempted to identify the type of microorganisms' changes that may enhance carcinogenesis (namely, sequencing advances are being crucial to understand how it happens) (9,10).
Therefore, the aim of this systematic review is to discuss and deepen possible changes in microbiota in adenoma-carcinoma cascade and its interaction with immune response.
Methods
Specific criteria were defined in order to guide this review. Firstly, a PubMed query to gather the articles related to the subject on was built: ("microbiota"[All Fields] OR "microbiome"[All Fields]) AND ("colorectal cancer"[All Fields] OR "colorectal carcinogenesis"[All Fields] OR "colon cancer"[All Fields] OR "rectal cancer"[All Fields]). With this query we intended to collect a wide range of articles, which then would be judiciously selected (total of 250 in November 2014).
A total amount of 254 articles were screened after the referred search (250 articles), cross-referencing (4 articles) and discarding the duplicates. The following inclusion criteria were used: a) Studies that were published until the end November 2014; b) the article should be written in English; and c) studies relevant to the subject (presenting original data). As exclusion criteria we defined: a) Studies considered by the authors as unrelated to the subject; and b) non-original studies. These criteria were applied by reading the title and abstract. After this step, 45 studies were selected for full-text reading. On a second level of eligibility, 14 more studies were excluded and 31 studies were selected, analysed and included in this revision (Table I and Fig. 1).
Results
Colorectal carcinogenesis
It is now believed there are two major pathways for colorectal carcinogenesis; the APC/β-catenin pathway (chromosomal instability) and the microsatellite instability pathway (associated with DNA mismatch repair genes).
The first pathway referred commonly appears as a consequence of mutations on oncogenes and tumor suppressor genes, and represents about 85% of colonic sporadic tumors. Firstly, APC tumor suppressor gene is lost; secondly, the other and intact allele of APC gene is also lost. Other mutations also occur, as in K-RAS, SMAD2, SMAD4 and p-53. Referring to the phenotype, colonic mucosa originates adenomas that become more dysplastic and may drive to carcinomas (11-13).
Further, in the second pathway (about 15% of sporadic colonic tumours) a precursor lesion might not be apparent. However, a serrated adenoma is believed to precede carcinoma. The inactivation of mismatch repair genes (mostly MLH1 and MSH2) is the main event in this cascade, and probably the first, conferring microsatellite instability. Microsatellites are mainly in noncoding regions, originating silent mutations; nevertheless, some are in the coding region or in promoting region of genes that regulate cell cycle and apoptosis (as TGF-β and BAX) (13-15).
Initial events leading to these mutations are still unknown. Subclinical inflammatory stimuli are potential initiators. For instance, our group has shown that colorectal carcinogenesis sequence is accompanied by an increase of Toll-like receptors (TLR) as well as its inhibitors lowering (like Toll-interacting protein, TOLLIP) in colonic lesions and that TLR2 and 4 polymorphisms strongly change colorectal carcinogenesis risk. Taking into account that animal models suggest that bacteria appear to be crucial to the development of colorectal cancer, it is thus hypothesized that the activation of the innate immunity receptors by bacteria leading to a chronic pro-inflammatory status may favor carcinogenesis (16-18).
Microbiota, inflammation and cancer
Microbiota in CRC patients: Microbial dysbiosis?
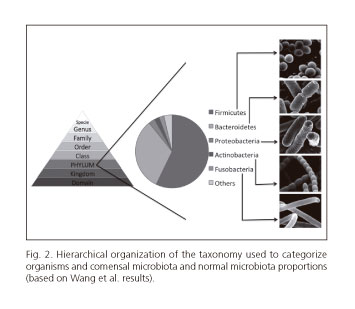
In order to correctly understand and study microbiota's change in colorectal carcinogenesis it is important to classify each bacterium. In general, organisms are categorized into hierarchic levels (19) (Fig. 2). In this review, microbiota is considered generally by phyla and when appropriated by genus and species.
Some of the phyla analysed below comprise gram-negative bacteria. In first place Fusobacteria is a phylum of anaerobic bacilli that may both be commensals or pathogens. Also, Bacteroidetes are anaerobic bacteria and are fully distributed in gastrointestinal tract. On its turn, Proteobacteria is a big phylum that houses more than 200 genera of gram-negative bacteria, including a wide variety of pathogens as E. coli, Salmonella and H. pylori (20).
On the other hand, other phyla include gram-positive bacteria. For example, Actinobacteria usually are aerobic and are frequently mistaken for fungi. Likewise, Firmicutes comprises mostly gram-positive bacteria (20).
It is also important to estimate the normal proportions of these bacteria. Besides the high variability found, the proportions of the most predominant bacteria are somehow consensual. The most prevalent bacteria are Firmicutes and Bacteroidetes (57.2% and 32.0% of colonic microbiota, respectively), according to Wang et al. These authors also stated that the second most predominant phyla seem to be Proteobacteria, Actinobacteria and Fusobacteria (representing, respectively, 2.81%, 2.22% and 2.20% of normal colonic microbiota) (21) (Fig. 2). In table II, we can see how these bacteria may change during colorectal carcinogenesis.
Fusobacteria
Fusobacterium, genus of this phylum, was shown to be more prevalent in colorectal cancer individuals than in healthy rats and humans (p = 0.001) (22,23). In addition, Kostic et al. found Fusobacterium in 48% of adenomas and, in those patients, it was augmented in adenomatous tissue vs. surrounding tissue - p < 0.004 (24). Furthermore, comparing individuals itself, the ones with higher abundance of Fusobacterium were apparently more likely to have adenomas (OR 3.66, 95% CI 1.37-9.74, p = 0.005) (25). This suggests these bacteria may start to accumulate early in the colorectal carcinogenesis sequence.
Moreover, Fusobacterium nucleatum (human's oral cavity colonizer), was found in a higher prevalence in CRC patient's faeces than in healthy individuals (60% vs. 22.2% respectively; p = 0.07) (26). It is still unknown whether this is a cause or a consequence (24,26).
Animal studies have shown that introducing human isolates of F. nucleatum in ApcMin/+ mouse model of intestinal tumorigenesis accelerates the onset of colonic tumours - the ones fed F. nucleatum developed a significantly higher number of colonic tumours compared to mice fed Streptococcus spp. (p < 0.001) (24).
Bacteroidetes
Referring to this phylum in general, a tendency to be augmented in the tumour, rather in adjacent mucosa, has been identified (27).
Bacteroides belong to this phylum and it is worth to mention that Wu et al. observed not only an increase of these bacteria in CRC but also a positive relation between the density of these bacteria and the disease status (R = 0.462, p = 0.046). Sobhani et al. that observed an increase from CRC patients stool and biopsy samples comparing with healthy individuals (5,22). This increase was also stated by animal-model based studies - for example, Zhu et al. shown that Bacteroides exhibited higher abundance in CRC rats compared with control animals (14.92% vs. 9.22%, p = 0.001). Also, Baxter et al., using transplanted faecal microbiota from both CRC patients and healthy individuals into germ-free mice, stated that Bacteroides where strongly correlated with increased tumor burden (p < 0.005). Finally, Zackular et al. found an enrichment of members of the Bacteroides in tumour-bearing mice (p < 0.001) (23,28,29).
Furthermore, Boleij et al. studied Enterotoxigenic Bacteroides fragilis (known for its role in acute diarrheal disease, inflammatory bowel disease, and colorectal cancer and which produce B. fragilis toxin) and concluded that the expression of the gene that encodes the toxin is strongly related with CRC (75% from CRC cases vs. 67% from healthy controls) and, particularly, with advanced CRC (100% from advanced stages vs. 72.7% from early stages; p = 0.093) (30).
Prevotella, genus which also belongs to Bacterioidetes phylum, was also shown to be overrepresented in CRC patients (p = 0.009) (5). Despite this, a mice-based study showed that family Prevotellaceae and, namely, members of the genus Prevotella were underrepresented in CRC animals (p < 0.05) (28).
Moreover, Alistipes (genus that belonging to the same phylum) were also in higher levels in tumour-bearing mice (relative abundance of 0.05) (29).
On the other hand, studies comparing Bacterioidetes levels between adenoma patients and controls, cases showed lower abundance of these bacteria (namely, showing lower proportions of Bacterioides spp.) - 29.14% vs. 34.14%, p < 0.05 (4). This result was also exhibited by Brim et al., in a study with pre-neoplastic lesions from African-Americans, particularly at a sub-genus level, and by animal studies in rats, comparing CRC animals with healthy ones (63.95% vs. 79.26%, respectively) (23,31). These results lead to hypothesise that Bacteroidetes may be fundamental at a cancer stage, rather than in adenoma-carcinoma sequence.
- Porphyromonadaceae: This is a bacteria family composed by genera as Porphyromonas and Dysgonomonas, which appear to be less prevalent in healthy individuals (p = 0.001) (22). In animal studies, Odoribacter (other genus of this family) was increased in CRC-mice supporting the previous statement; though, different results were found for other members of this family, which appeared to be underrepresented in CRC-mice (28). Additionally, Parabacteroides (another genus of this family) were also shown to be positively related to tumorigenesis rate in mice transplanted with faecal microbiota from CRC patients, comparing with the ones transplanted with healthy individuals microbiota (29).
Actinobacteria
Bifidobacterium represents a ubiquitous, gastrointestinal, vaginal and oral cavity colonizer gram-positive bacterium, belonging to Actinobacteria phylum; it seems to be higher in control individuals than in CRC-patients (26). Also, when comparing colon cancer patients with diverticulitis patients, the first have lower counts of Bifidobacterium (Bifidobacteria were found in 100% of patients with diverticulitis and in 76% of those suffering colon cancer) (26,32).
Coriobacteridae is a subclass which has been demonstrated to be increased in CRC tissue, namely the genera Slackia and Collinsella, regarded as gastrointestinal commensals (27).
Firmicutes
This is one of the predominant phyla both in health and disease. Although, when looking to the whole phylum, there is no difference between adenoma cases and healthy controls, specific subgroups show differences (4).
At a family level, Staphylococcaceae was shown to be higher in CRC patients than in healthy controls (p = 0.011) (22).
On the other hand, Clostridiales are a class of gram-positive bacteria that were negatively correlated with CRC formation in a mice-based study, which used faecal transplantation (29).
More specific studies on species from this class have been done; namely intraindividual temporal stability of Clostridium coccoides, is significantly (p < 0.05) different comparing CRC patients (65%) with both healthy controls (76%) and polypectomized individuals (77%). Furthermore, C. leptum's temporal stability was also lower (although not significant) in CRC patients (33).
Eubacteriaceae, on its turn, is a family which belongs to Clostridiales order and that is statistically significant augmented in CRC patients (p = 0.037) (22). Despite this, Eubacterium, a butyrate-producing genus of this family, was reduced in CRC rats, comparing with the control group (23).
Lactobacillus, is a probiotic specie which also belongs to this phylum, seems to be diminished (although not statistically significant) in faecal samples of tumour patients rather than in healthy controls (p = 0.064). This decrease in CRC patients (comparing with healthy controls) was also observed in animal studies with rats (2.32% vs. 3.71%; p < 0.001). Furthermore, Ruminococcus, another probiotic specie, was similarly decreased (23,26).
- Faecalibacterium prausnitzii: Bacteria generally present as a commensal in gastrointestinal tract and which was demonstrated to be diminished in CRC patients (13.3% vs. 40%; p = 0.06) (22,26). Even though, different results are found comparing patients with adenomas with control individuals; Shen et al. found that Faecalibacterium spp. were increased in case subjects (21.7% vs. 10.3%, p < 0.05) (4).
Roseburia, which is a butyrate-producing genus, as well as Faecalibacterium, were also diminished in CRC patients (3.59% in healthy controls and 1.56% in CRC patients; p < 0.05); the same tendency was also seen in rats by Zhu et al. (21-23).
Proteobacteria
Analysing at a phylum level, components of this one were shown to be overrepresented in adenoma cases, comparing with controls (12.9% vs. 4.85%, respectively; p < 0.05) (4).
Animal studies in rats (animal model of 1,2-dimethylhydrazine - induced colon cancer) show no difference in the species of Proteobacteria between CRC animals and healthy controls, when comparing bacterial communities (p = 0.175), although its abundance was higher in CRC animals (1.06% in healthy rats, comparing with 2.95% in CRC rats) (23).
Enterobacteriaceae is a family from this phylum. It includes both harmless microorganisms and pathogens, and it seems to be augmented in cancer patients (46.6% in CRC patients vs. 0% in control group). In fact, when splitting tubular adenoma and adenocarcinoma patient's results, the last one has statistically significant higher levels of these bacteria (p = 0.035), which may lead to the hypothesis that there may be a positive correlation between bacteria level and tumour's stage (26). Nonetheless, controversial results were found; actually, members of this family, as Citrobacter, Shigella, Cronobacter, Kluyvera, Serratia and Salmonella spp., have been shown to be lessened in cancer tissue, comparing with adjacent mucosa (27).
Further, Campylobacteraceae, other family from this phylum, has been shown to be less prevalent in healthy controls (p = 0.014) (22).
- E. coli: E. coli (which belongs to Proteobacteria phylum) represents gram-negative commensal bacterium from human gut, although some strains have been recognized as pathogens, associated with an inflammation status and toxin production (as cyclomodulin). Studies have demonstrated that cyclomodulin-positive pathogenic strains are more prevalent in most advanced cancer stages (using TNM staging, TNM I - 45%; TNM II - 64% and TNM III/IV - 67%); furthermore, there is evidence of an increase of mucosa-associated and internalized E. coli in tumour's tissue, comparing with normal mucosa (from diverticulitis patients' controls). These two were also significantly correlated with proliferative index of the mucosa (p < 0.02 and p < 0.04, respectively) (34).
Still, experiments with IL-10-/- mice showed that host's inflammation is needed for E. coli cancer-promoting activity: In the absence of inflammation, high abundance of E. coli was not sufficient for tumorigenesis (35).
Other bacteria
- Treponema. Zhu et al. showed that, in rats, these gram-negative bacteria, belonging to Spirochaetes phylum, were reduced among CRC-rats group (2.43% in CRC group vs. 3.04% in control group, p < 0.001) (23).
- Methanobacteriales. This represents a group from Euryarchaeota phylum. Methanobacteriales were higher in polyps and tumours, rather than in healthy controls. Actually, a positive correlation was seen between CRC and Methanobacteriales presence (r = 0.537, p = 0.007). To be more accurate, significantly higher levels of faecal bacteria were observed when comparing the control group with the CRC group (p = 0.0033); however, no differences between controls and adenoma cases were observed (p = 0.48). Moreover, no significant differences between tumour and adenoma samples were found (p = 0.189) (26).
- Akkermansia muciniphila. Is an anaerobic, gram-negative, mucin-degrading bacterium (Verrucomibrobia phylum), which appears to be in higher levels in tumour patients (33.3% vs. 0%; p = 0.136 according to Mira-Pascual et al.; and 12.8% vs. 3.54%, according to Weir et al.) (26,36). Akkermansia genus was also elevated in tumour-induced mice, rather than in healthy ones (28,29).
Bacterial metabolism
Butyrate
Butyrate is thought to protect against colonic inflammation and, therefore, CRC (36,37). Butyrate-producing bacteria seem to be underrepresented in CRC patients. Studies reveal that faeces of healthy individuals show higher butyrate levels, comparing with acetate rich CRC-patients stool (29,36).
GPR109a, coded by Niacr1, is a receptor for this metabolite and for niacin (metabolite that prevents inflammation). It's signalling stimulates anti-inflammatory response, allowing the differentiation of anti-inflammatory cells (namely regulatory T cells and IL-10 producing cells). Experiments with Niacr1-/- mice have shown that those were more susceptible to colonic cancer. This may be a good starting point for new prevention and therapeutic measures using, for example, a receptor agonist (37).
Other fatty acids
Healthy controls presented higher levels of poly and monounsaturated fatty acids in stool than CRC-cases. Healthy controls also presented higher ursodeoxycholic acid and glycerol (glycerol might be taken up by tumour cells) levels (36).
Amino acids
Faeces from CRC-patients show higher concentrations of amino acids; this might be due to a great variety of reasons as differences in protein consumption by the different microorganisms, reduction in nutrient's absorption because of the inflammation status and/or augmented autophagy, among others (36,38).
Cyclomodulin
Cyclomodulin, toxin produced by some (pathogenic) strains of E. coli, may participate in the carcinogenesis pathway. It is genotoxic and has great impact in several cell functions (cell-cycle progression, proliferation, differentiation, apoptosis, etc.). A particular experiment with E. coli strain 11G5, that encodes colibactin (which acts as a cyclomodulin), revealed that mice colonized with it showed greater tumor's size and number (34).
Actually, this toxin was higher in adjacent normal mucosa of colonic cancer patients, rather than in the diverticulitis ones (the colonic mucosa of CRC patients was mostly colonized by B2 cyclomodulin-producing phylogroup); moreover, this strains were also augmented in higher tumour's stages, comparing with the lower ones (34,39).
This information suggests that the association between the microbiota and CRC may be due to metabolic activity (Table III), overlapping bacterial phylogeny.
Biomarkers
Latest studies have emphasized the possibility of creating a new, non-invasive diagnostic test of CRC, using biomarkers (in fecal material).
Zackular et al. combined the characterization of microbiome in healthy, adenoma and carcinoma patients with recognized risk factors for CRC, and concluded that the discrimination between these groups strongly ameliorated, rather than the risk factors alone (40).
Analysing and investigating the presence of bacterial metabolites, new CRC biomarkers may arise, using a non-invasive diagnostic test (feces samples).
Further research is needed to test and consolidate these hypotheses and also to test the use of bacteria and various metabolites in order to create a new, non-invasive and trustable diagnostic test which can benefit clinical practise.
Immune signalling and inflammation
The impact of microbiota changes in immune signalling is undeniable and so it is the role of chronic inflammation in colorectal carcinogenesis. There are many published results that brace this idea.
McCoy et al. showed a significant positive correlation in CRC cases between Fusobacterium prevalence and local inflammatory cytokines gene expression, explicitly TNF-α (r = 0.33, p = 0.06) - suggesting that Fusobacterium may increase mucosal inflammation- and, remarkably, IL-10 (r = 0.44, p = 0.01), highlighting the complexity of the relationship host/intestinal microbiota (25).
When comparing healthy individuals' mucosa with adjacent mucosa of tumour lesion from CRC patients, the latter showed overrepresentation of IL-17 immunoreactive cells (mostly CD3 marking cells). Interestingly, not only IL-17A, but also IL-17C have been proved to be up regulated in CRC, both in human and mouse models, although being differentially regulated (5,41).
Further investigations on IL-17C production have led to new knowledge on this subject; even though it is up regulated in CRC, IL-17C signalling is crucial for and promotes tumorigenesis (by the induction of Bcl-2 and Bcl-xL expression). Moreover, throughout tumorigenesis microbiota drives its production leading, therefore, to CRC promotion (41).
Notwithstanding, a different investigation on IL-10 showed that its deficiency in mice worsened epithelial mitosis and polyp's growth; also IL-10-/- mice had more, more penetrant and multiple tumours. In addition, the expression of IL-12p40 and TNF-α mRNA was shown to be strongly augmented in IL-10-/- mice (18,42).
TLR/MyoD88 was also shown to be crucial in bacterial-induced carcinogenesis (inflammation-related CRC); in fact, IL-10-/-; MyoD88-/- mice lacked neoplastic signs, unlike IL-10-/- mice. On the other hand, IRAK-M (IL-1 receptor associated kinase-M) deficient mice presented invasive and antibiotic resistant cancer (18,43).
Further human research showed changes on TLR's profile in normal mucosa adjacent to the lesion (either adenoma or carcinoma), comparing with normal, healthy subjects' mucosa. This supports TLR's involvement in CRC. Namely, a persistently positive TLRs expression (as TLR2) and lower expression of TLRs inhibitors (as TOLLIP) were associated with higher TLRs protein levels throughout all the spectrum of lesions of colon carcinogenesis (16).
TGF-beta signalling pathway is also important in gut's inflammatory and microbiota microenvironment. It is crucial down-regulating the inflammatory response a maintaining the normal gut environment. Actually, SMAD4's (mediator of this pathway) haploinsufficiency was reported in two case reports and, then, tested in mice. The resultant inflammatory environment may be connected with CRC development (44).
Supplementary information shows that c-Jun/JNK and STAT3 signalling pathways are generated by colonic microbiota and anaemia, respectively and that both are thought to act synergistically in tumour's growth in APCMin/+ mice. Supporting this statement it was seen that germ-free APCMin/+ practically lacked colonic tumours (45).
Nonetheless, some investigations have been made on inflamossomes, cytoplasmic protein complexes (Nod-like receptors- NLR's, pro-caspase-1 and eventually the adaptor protein apoptosis-associated speck-like protein containing a CARD- ASC). ASC-/- mice showed greater predisposition for CRC tumorigenesis, as well as NLRP6-/-mice. Besides, intriguingly, wild-type mice cohoused with ASC-/- and NLRP6-/- mice had a great increase in the propensity to develop an inflammation-induced CRC, comparing with singly housed ones- can this represent a transmissible cancer? (46).
Discussion
The information resulting from the revised studies suggest that several different bacteria, isolated or acting concurrently, may play an important role in colorectal carcinogenesis. Evidence was gathered that this may happen by proinflammatory and metabolic stimulus. It looks clear that investigation of colon microbiota and its cellular pathways is an area of great potential.
However, several limitations must be assigned to this review and to the studies herein included. Firstly, it both includes human and animal studies, giving similar importance to both. Furthermore, different authors used different methodology (namely the usage of fecal samples/biopsy samples; the usage of biopsies from normal adjacent mucosa compared with tumors' mucosa versus normal healthy mucosa compared with tumors' mucosa; number of patients/animals used, etc.), which may affect the achieved results and can contribute to the observed differences. Despite the inherent limitations, the present review allowed us to reach valid and important results.
It seems clear that colorectal carcinogenesis is associated with a microbial dysbiosis. Actually, although with some controversial results, in carcinoma patients (either humans or animals) some bacteria were consistently found augmented (such as Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteridae, Staphylococcaceae, Akkermansia spp. and Methanobacteriales), while other were constantly diminished (such as Bifidobacterium, probiotic species- namely Lactobacillus and Ruminococcus, butyrate-producing bacteria- explicitly Faecalibacterium spp., Roseburia-, and Treponema).
Concerning to bacterial metabolites, it looks sharply defined that butyrate-producing bacteria are lessened in CRC patients, as well as poly and monounsaturated fatty acids and ursodeoxycholic acid (29,36). Also, higher concentrations of amino acids are found in CRC individuals (36,38). Notwithstanding, cyclomodulin-producing E. coli strains were augmented in higher tumour's stages, comparing with the lower ones (34,39). All this evidence hints that metabolic environment might be deeply involved in colorectal carcinogenesis.
Moreover, the gathered information leads to the conclusion that different microbiota may have different effects in immune signalling and that this may contribute to a chronic proinflammatory stimulus in colorectal carcinogenesis. The results suggest that dysbiosis may be the missing link between the several studies that show immunologic changes in the colon mucosa (cytokine profile, TLR's expression) and colorectal cancer. Definitely, microbiota changes are closely related to inflammatory environment and to CRC's development. Other reviews on the subject also support this statement (47-50).
In view of this information, it seems appropriate to hypothesize that different bacterial profiles exposes colonic mucosa to different metabolic (given that different bacteria originates different metabolites) and inflammatory stimuli (namely, by activation of the innate immune response), and that this seems to create a subclinical pro-inflammatory state that enables DNA mutations and, therefore, colorectal carcinogenesis (Fig. 3).
Taking altogether, and even though from the analysis of the studies included in this review it is not possible to firmly conclude that dysbiosis is a cause or a consequence in CRC, it looks clear that future research in this area is needed. Moreover, methods to study colonic microbiota should be homogenised in order to compare studies and to reach valid conclusions. The precise analysis of CRC microenvironment (composition, immunologic pathways and metabolite production) seems to deserve to be seen as a major point of interest in scientific research nowadays. Dietary and drug (possibly using antibiotics, probiotics and anti-inflammatory drugs) approaches might well be tried and further adopted in clinical practise. Furthermore, the usage of metabolites appears to be very useful as a non-invasive way of diagnosing this pathology.
In conclusion, microbiota appears to have an important role in colorectal carcinogenesis. However, more studies applying validated techniques are needed. It is predictable that in the future new prophylactic, diagnostic and therapeutic strategies can arise from microbiome research.
References
1. Candela M, Guidotti M, Fabbri A, et al. Human intestinal microbiota: Cross-talk with the host and its potential role in colorectal cancer. Crit Rev Microbiol 2011;37:1-14. DOI: 10.3109/1040841X.2010.501760. [ Links ]
2. Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science 2006;312:1355-9. DOI: 10.1126/science.1124234. [ Links ]
3. Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature 2007;449:804-10. DOI: 10.1038/nature06244. [ Links ]
4. Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 2010;1:138-47. DOI: 10.4161/gmic.1.3.12360. [ Links ]
5. Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One 2011;6:e16393. DOI: 10.1371/journal.pone.0016393. [ Links ]
6. Rustgi AK. The genetics of hereditary colon cancer. Genes & development. 2007;21:2525-38. DOI: 10.1101/gad.1593107. [ Links ]
7. Watson AJ, Collins PD. Colon cancer: A civilization disorder. Dig Dis. 2011;29:222-8. DOI: 10.1159/000323926. [ Links ]
8. Riscuta G, Dumitrescu RG. Nutrigenomics: Implications for breast and colon cancer prevention. Methods Mol Biol 2012;863:343-58. [ Links ]
9. Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541-6. DOI: 10.1038/nature12506. [ Links ]
10. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. DOI: 10.1126/science.1235122. [ Links ]
11. Groden J, Thliveris A, Samowitz W, et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 1991;66:589-600. DOI: 10.1016/0092-8674(81)90021-0. [ Links ]
12. Medema JP, Vermeulen L. Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature 2011;474:318-26. DOI: 10.1038/nature10212. [ Links ]
13. Cunningham D, Atkin W, Lenz HJ, et al. Colorectal cancer. Lancet 2010;375:1030-47. DOI: 10.1016/S0140-6736(10)60353-4. [ Links ]
14. Poulogiannis G, Frayling IM, Arends MJ. DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 2010;56:167-79. DOI: 10.1111/j.1365-2559.2009.03392.x. [ Links ]
15. Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol 2003;21:1174-9. DOI: 10.1200/JCO.2003.04.060. [ Links ]
16. Pimentel-Nunes P, Goncalves N, Boal-Carvalho I, et al. Decreased Toll-interacting protein and peroxisome proliferator-activated receptor gamma are associated with increased expression of Toll-like receptors in colon carcinogenesis. Journal of Clinical Pathology 2012;65:302-8. DOI: 10.1136/jclinpath-2011-200567. [ Links ]
17. Pimentel-Nunes P, Teixeira AL, Pereira C, et al. Functional polymorphisms of Toll-like receptors 2 and 4 alter the risk for colorectal carcinoma in Europeans. Digestive and Liver Disease 2013;45:63-9. DOI: 10.1016/j.dld.2012.08.006. [ Links ]
18. Uronis JM, Muhlbauer M, Herfarth HH, et al. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One 2009;4:e6026. DOI: 10.1371/journal.pone.0006026. [ Links ]
19. Tyler AD, Smith MI, Silverberg MS. Analyzing the human microbiome: A "how to" guide for physicians. The American Journal of Gastroenterology. 2014;109:983-93. DOI: 10.1038/ajg.2014.73. [ Links ]
20. Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology 2007;88:1354-64. DOI: 10.1890/05-1839. [ Links ]
21. Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME Journal 2012;6:320-9. DOI: 10.1038/ismej.2011.109. [ Links ]
22. Wu N, Yang X, Zhang R, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol 2013;66:462-70. DOI: 10.1007/s00248-013-0245-9. [ Links ]
23. Zhu Q, Jin Z, Wu W, et al. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLoS One 2014;9:e90849. DOI: 10.1371/journal.pone.0090849. [ Links ]
24. Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013;14:207-15. DOI: 10.1016/j.chom.2013.07.007. [ Links ]
25. McCoy AN, Araujo-Perez F, Azcarate-Peril A, et al. Fusobacterium is associated with colorectal adenomas. PLoS One 2013;8:e53653. DOI: 10.1371/journal.pone.0053653. [ Links ]
26. Mira-Pascual L, Cabrera-Rubio R, Ocon S, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol 2014;10.1007/s00535-014-0963-x. DOI: 10.1007/s00535-014-0963-x. [ Links ]
27. Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS One 2011;6(5):e20447. DOI: 10.1371/journal.pone.0020447. [ Links ]
28. Zackular JP, Baxter NT, Iverson KD, et al. The gut microbiome modulates colon tumorigenesis. MBio 2013;4:e00692-13. DOI: 10.1128/mBio.00692-13. [ Links ]
29. Baxter NT, Zackular JP, Chen GY, et al. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2014;2:20. DOI: 10.1186/2049-2618-2-20. [ Links ]
30. Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis 2015;60:208-15. DOI: 10.1093/cid/ciu787. [ Links ]
31. Brim H, Yooseph S, Zoetendal EG, et al. Microbiome analysis of stool samples from African Americans with colon polyps. PLoS One 2013;8:e81352. DOI: 10.1371/journal.pone.0081352. [ Links ]
32. Gueimonde M, Ouwehand A, Huhtinen H, et al. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J Gastroenterol 2007;13:3985-9. DOI: 10.3748/wjg.v13.i29.3985. [ Links ]
33. Scanlan PD, Shanahan F, Clune Y, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environmental Microbiology 2008;10:789-98. DOI: 10.1111/j.1462-2920.2007.01503.x. [ Links ]
34. Bonnet M, Buc E, Sauvanet P, et al. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res 2014;20:859-67. DOI: 10.1158/1078-0432.CCR-13-1343. [ Links ]
35. Arthur JC, Gharaibeh RZ, Muhlbauer M, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun 2014;5:4724. DOI: 10.1038/ncomms5724. [ Links ]
36. Weir TL, Manter DK, Sheflin AM, et al. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 2013;8:e70803. DOI: 10.1371/journal.pone.0070803. [ Links ]
37. Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014;40:128-39. DOI: 10.1016/j.immuni.2013.12.007. [ Links ]
38. Sato K, Tsuchihara K, Fujii S, et al. Autophagy is activated in colorectal cancer cells and contributes to the tolerance to nutrient deprivation. Cancer Res 2007;67:9677-84. DOI: 10.1158/0008-5472.CAN-07-1462. [ Links ]
39. Buc E, Dubois D, Sauvanet P, et al. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One 2013;8:e56964. DOI: 10.1371/journal.pone.0056964. [ Links ]
40. Zackular JP, Rogers MA, Ruffin MTt, et al. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila). Cancer Prev Res (Phila) 2014;10.1158/1940-6207.capr-14-0129. DOI: 10.1158/1940-6207.CAPR-14-0129. [ Links ]
41. Song X, Gao H, Lin Y, et al. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity 2014;40:140-52. DOI: 10.1016/j.immuni.2013.11.018. [ Links ]
42. Dennis KL, Wang Y, Blatner NR, et al. Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Cancer Res 2013;73:5905-13. DOI: 10.1158/0008-5472.CAN-13-1511. [ Links ]
43. Klimesova K, Kverka M, Zakostelska Z, et al. Altered gut microbiota promotes colitis-associated cancer in IL-1 receptor-associated kinase M-deficient mice. Inflamm Bowel Dis 2013;19:1266-77. DOI: 10.1097/MIB.0b013e318281330a. [ Links ]
44. Szigeti R, Pangas SA, Nagy-Szakal D, et al. SMAD4 haploinsufficiency associates with augmented colonic inflammation in select humans and mice. Ann Clin Lab Sci 2012;42:401-8. [ Links ]
45. Li Y, Kundu P, Seow SW, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis 2012;33:1231-8. DOI: 10.1093/carcin/bgs137. [ Links ]
46. Hu B, Elinav E, Huber S, et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc Natl Acad Sci U S A 2013;110:9862-7. DOI: 10.1073/pnas.1307575110. [ Links ]
47. Irrazabal T, Belcheva A, Girardin SE, et al. The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell 2014;54:309-20. DOI: 10.1016/j.molcel.2014.03.039. [ Links ]
48. Turner ND, Ritchie LE, Bresalier RS, et al. The microbiome and colorectal neoplasia: Environmental modifiers of dysbiosis. Curr Gastroenterol Rep 2013;15:346. DOI: 10.1007/s11894-013-0346-0. [ Links ]
49. Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317-28. DOI: 10.1016/j.chom.2014.02.007. [ Links ]
50. Hagland HR, Soreide K. Cellular metabolism in colorectal carcinogenesis: Influence of lifestyle, gut microbiome and metabolic pathways. Cancer Lett 2014 Mar 12;10.1016/j.canlet.2014.02.026. [ Links ]
![]() Correspondence:
Correspondence:
Marta Borges-Canha.
Department of Physiology and Cardiothoracic Surgery,
Cardiovascular Research & Development Unit.
Faculty of Medicine.
Al. Prof. Hernâni Monteiro,
4200-319, Porto, Portugal
e-mail: marta.canha@gmail.com
Received: 29-04-2015
Accepted: 29-07-2015