Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Revista Española de Enfermedades Digestivas
versión impresa ISSN 1130-0108
Rev. esp. enferm. dig. vol.109 no.12 Madrid dic. 2017
https://dx.doi.org/10.17235/reed.2017.4426/2016
Methylation status of the estrogen receptor 1 promoter predicts poor prognosis of acute-on-chronic hepatitis B liver failure
Xiao-Peng Fan1, Cheng-Yun Dou1, Yu-Chen Fan1,2, Chuang-Jie Cao3, Ze-Hua Zhao1 and Kai Wang1,2
1Department of Hepatology. Qilu Hospital of Shandong University, Jinan, China.
2Hepatology Institute of Shandong University. Jinan, China.
3Department of Pathology. The First Affiliated Hospital of Sun Yat-san University. Guangzhou, China
Contributed equally: Xiao-Peng Fan and Cheng-Yun Dou.
This work was supported by grants from the Science and Technology Development Plan of the Shandong Province (2014GSF118068), the National Natural Science Foundation of China (81171579, 81201287, 81371832) and the Key Project of the Chinese Ministry of Science and Technology (2012ZX10002007, 2013ZX10002001).
ABSTRACT
Background: Acute-on-chronic hepatitis B liver failure (ACHBLF) is an acute deteriorating liver disease and rapidly progresses to multiple organ failure. There is currently no adequate accurate predictive models of ACHBLF prognosis.
Aims: To identify the methylation frequency of the estrogen receptor 1 (ESR1) promoter in ACHBLF and analyze the associated prognostic significance.
Methods: Methylation-specific PCR (MSP) was used to determine the methylation frequency of the ESR1 promoter in peripheral blood mononuclear cells from a training and validation cohort of patients. The training cohort included 113 patients with ACHBLF, 73 with chronic hepatitis B (CHB) and 40 healthy controls (HCs). The validation cohort consisted of 37 patients with ACHBLF. Another 18 patients with pre-ACHBLF who progressed to ACHBLF were used to dynamically evaluate ESR1 promoter methylation changes associated with a severe clinical condition.
Results: Death from ACHBLF was associated with hyperbilirubinemia, a higher score in the model for end-stage liver disease (MELD), a higher incidence of hepatic encephalopathy (HE) and an increased frequency of ESR1 promoter methylation during the 28 day follow-up. HE, MELD score and ESR1 promoter methylation were the independent risk factors associated with 28-day mortality from ACHBLF. The frequency of ESR1 promoter methylation was significantly higher than in patients with CHB and HCs. Albumin and the MELD score were significantly associated with ESR1 promoter methylation. Moreover, ESR1 promoter methylation frequency increased with ACHBLF progression. More importantly, ESR1 promoter methylation was an independent risk factor and had a high value to predict 28-day mortality from ACHBLF.
Conclusions: Abnormal ESR1 methylation could be a prognostic biomarker for ACHBLF.
Key words: Acute-on-chronic hepatitis B liver failure. ESR1. Methylation. Prognostic biomarker.
Introduction
Acute-on chronic liver failure (ACLF) is defined as an acute and severe hepatic insult based on an underlying chronic liver disease or liver cirrhosis (1). Hepatitis B virus (HBV) associated ACLF, also known as acute-on-chronic hepatitis B liver failure (ACHBLF), remains the predominant ACLF in the Asian area, especially in China (2,3). ACHBLF progresses rapidly and the mortality rate is between 50% and 90% (4). Liver transplantation is the most effective treatment. However, due to a shortage of liver donors, transplantation is performed in fewer than 10% of ACHBLF patients worldwide (5). The model for end-stage liver disease (MELD) is the most commonly used prediction model to forecast patient survival, but it does not take into account HBV infection, ascites and hepatic encephalopathy (HE) (6). Therefore, an objective and accurate predictive model, which can identify patients with a high probability of mortality is needed in order to prioritize liver transplantation cases and increase survival of patients with ACHBLF.
Estrogens are endogenous antioxidants and play a major role in attenuating hepatocyte apoptosis and suppressing hepatic fibrosis (7). In addition, estrogens play an anti-carcinogenic role in patients with chronic hepatitis (8). Estrogens exert their effects primarily via the estrogen receptor 1 (ESR1) in the liver (9). Aberrant ESR1 expression in the liver has been implicated in the stimulation of hepatocyte injury and may act as an inducer of liver disease (10), including chronic alcohol liver disease (11), benign hepatic neoplastic disease (12) and hepatocellular carcinoma (HCC) (13). Furthermore, the frequency of abnormal ESR1 expression was higher in male patients than in females, even at an early stage of chronic liver disease (14). HBV, is a sex hormone response virus and the liver is a sexually dimorphic organ (15). Regardless of pathophysiology, ACHBLF is a complex autoimmune disease with a higher prevalence in men than in women. This gender disparity may be due to a lower production of estrogen and/or a reduced response to estrogen. Due to the fact that HBV-related HCC occurs more often in men than in women and ESR1 promoter methylation exists in this disease (16), it is possible that methylation of the ESR1 promoter may play an important role in patients with ACHBLF.
Deoxyribonucleic acid (DNA) methylation of specific genes as a clinical biomarker has been largely explored in various diseases (17,18). Methylation modification is a mechanism contributing to the downregulation of ESR1 gene expression (19). Numerous studies have reported the association of ESR1 promoter methylation with HCC (20), breast cancer (21) and endometrial cancer (22). After HBV DNA integrates into the human genome, HBV DNA and the encoded protein hepatitis Bx (HBx) increases the expression of DNA methyltransferases inducing gene promoter hypermethylation and the downregulation of gene expression (23). Therefore, ESR1 promoter hypermethylation may be caused by HBV infection. However, there are few studies that evaluate the potential role of ESR1 methylation in the progression of ACHBLF. Peripheral blood monocytes (PBMCs) travel throughout the body and respond to various external and internal stimuli (24). These cells are exposed to various metabolically relevant tissues/organs including the liver, adipose tissue and endothelium, and are also known to cross-talk with these organs (25). Moreover, PBMCs and hepatic cells are similar with regard to several functions (26). Therefore, PBMCs could serve as a less invasive and direct alternative to liver tissue. In this study, methylation-specific polymerase chain reaction (MSP) was performed to determine the methylation frequency of the ESR1 promoter from PBMCs in a training cohort and a validation cohort that consisted of 37 patients with ACHBLF. The association of ESR1 promoter methylation with clinical characteristics was investigated and the potential predictive value of ESR1 promoter methylation for 28-day mortality in patients with ACHBLF was analyzed.
Patients and methods
Study population
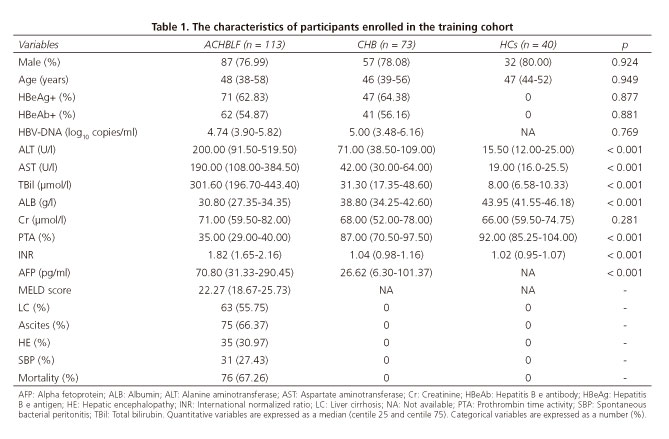
A total of 168 patients with ACHBLF, 73 patients with chronic hepatitis B (CHB) and 40 healthy controls (HCs) were recruited from the Department of Hepatology of the Qilu Hospital of Shandong University from September 2009 to January 2016. Of the 168 ACHBLF patients, 113 patients hospitalized from September 2009 to August 2014 were defined as the training cohort and 37 patients hospitalized from September 2014 to January 2016 were defined as the validation cohort. The remaining 18 patients were initially pre-ACHBLF and eventually progressed to ACHBLF. The diagnosis of ACHBLF was made according to the 2009 guidelines (27). All patients with ACHBLF had acute hepatic insult manifesting as jaundice (serum total bilirubin [TBil] ≥ 85 μmol/l) and coagulopathy (international normalized ratio [INR] ≥ 1.5 or prothrombin activity [PTA] < 40%), with complications arising within four weeks due to ascites and/or encephalopathy in a patient with a previous diagnosis of CHB. Pre-ACHBLF was defined as 1.28 ≤ INR < 1.5 or 40% < PTA ≤ 60% (28). According to the update for the management of CHB, this is defined as a positive B surface antigen for at least six months prior to the beginning of this study (29). Liver cirrhosis was either proven histopathologically or by explicit morphological criteria of liver cirrhosis with ultrasound, computed tomography or magnetic resonance imaging (30).
All patients were newly diagnosed and had not received any treatment. Patients with hepatitis C, hepatitis D or human immunodeficiency virus infection and any other type of liver disease were excluded. Informed consent was obtained from each participant before fasting blood samples were collected on the second day of hospitalization. The Medical Ethical Committee in the Qilu Hospital of Shandong University approved the study. The patient selection process is shown in figure 1.
The first day of follow-up was the day of diagnosis of ACHBLF. All patients with ACHBLF were followed up for 28 days and the outcome was recorded (non-survival or survival). None of the patients included in the study underwent liver transplantation.
Clinical and laboratory parameters
Hepatitis B surface antigen (HBsAg), hepatitis B e antigen (HBeAg), and hepatitis B e antibody (HBeAb) were detected by an electrochemiluminescence assay (Roche Diagnostic GmbH, Mannheim, Germany). Laboratory tests of liver function, such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), total bilirubin (TBil), albumin (ALB) and coagulation function were performed using the Roche automatic biochemical analyzer Cobas c311 (Roche Diagnostic GmbH). MELD scores were calculated on admission according to the original formula proposed by the Mayo Clinic group: MELD score = 3.78 × LN (bilirubin [mg/dl]) + 11.2 × LN (INR) + 9.57 × LN (creatinine [Cr; mg/dl]) + 6.4 × (etiology: 0 if cholestatic or alcoholic, 1 for otherwise) (31).
Peripheral blood mononuclear cells isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from 5 ml of whole blood via a gradient centrifugation method (1,500 rpm × 5 min → 2,000 rpm × 20 min → 2,000 rpm × 10 min → 3,000 rpm × 5 min) using Ficoll-Paque plus density gradient medium (GE Healthcare, Uppsala, Sweden) according to the manufacturer's instructions. Cells were harvested and stored at -20 oC for later use.
RNA and cDNA preparation from PBMC and quantitative real-time PCR
Total RNA of PBMC was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following a conventional procedure. Two micrograms of total RNA from each sample were reverse transcribed into cDNA using the ReverAid First Strand cDNA Synthesis kit (Fermentas, Vilnius, Lithuania). The expression of ESR1 mRNA was evaluated by qPCR in triplicate. GAPDH was used as the endogenous control to normalize RNA quantity and quality differences between samples. Primer sequences were as follows: ESR1 forward: 5'- CACCAGATCCAAGGGAA -3' and reverse: 5'- CGGCGTTGAACTCGTAG -3'; GAPDH forward: 5'-GACTCATGACCACAGTCCATGC-3' and reverse: 5'- AGAGGCAGGGATGATTGTTCTG-3'. The qPCR assay was performed with 75 ng of template cDNA, 10 μl 2 × SYBR Green Premix Ex Taq™ (Takara, Japan), forward primer 0.5 μl (10 μM), reverse primer 0.5 μl (10 μM) and DNase/RNase-free H2O up to a total volume of 20 μl using the Lightcycler 480 (Roche Diagnostics, Germany) according to the manufacturer's protocol. The PCR protocol was 95 oC for 30 sec, followed by 40 cycles of 95 oC for 5 sec, 60 oC for 30 sec and 72 oC for 30 sec. A melting-curve analysis was performed to ensure the specificity of gene amplifications. All PCR product quantities were determined using the comparative method.
DNA purification from PBMC and bisulfite conversion
Total genomic DNA was extracted with sodium citrate-ethanol-sodium hydroxide (NaOH) from PBMC residue after extracting RNA with the Trizol reagent (Invitrogen, Carlsbad, CA, USA). DNA was dissolved in 8 mM NaOH and the concentration obtained was between 200-300 ng/μl.
DNA bisulfite conversion was strictly carried out according to the instruction manuals of the Zymo Research EZ DNA Methylation-Gold™ Kit (Zymo Research, Orange, CA, USA). In order to ensure optimal results, the amount of input DNA was 500 ng. Excessive DNA input may result in incomplete template cytosine to uracil conversion and affect the outcome of MSP. Depending on the requirements of the experiment, the final bisulfite-modified DNA pellets were dissolved at 20 μl M-Elution Buffer and stored at -80 oC.
Analysis of DNA methylation of ESR1 promoter by MSP
Bisulfite-modified DNA was amplified with primers specific for methylated (M) or unmethylated (U) sequences by MSP. Methylated forward primer: 5'-ACGAGTTTAACGTCGCGGTC-3' and reverse: 5'-ACCCCCCAAACCGTTAAAAC-3'; and unmethylated forward primer: 5'-TGTTGTTTATGAGTTTAATGTTGTGGTT-3' and reverse: 5'-AAAAAAACCCCCCAAACCATT -3'. Amplification was carried out in a 25-μl reaction mixture containing 1 μl bisulfite-modified DNA, ZymoTaq™ premix 12.5 μl, 0.5 μl of forward primer (10 μM), 0.5 μl of reverse primer (10 μM) and 10.5 μl of DNase/RNase-free H2O (ZymoTaq™ DNA polymerase; Zymo Research). The PCR protocol was performed at an initial denaturation at 95 oC for 10 min, followed by 40 cycles of denaturation at 95 oC for 30 sec, annealing at 58 oC for 40 sec, extension at 72 oC for 40 sec and final extension at 72 oC for 10 min. The PCR products were confirmed on 2% agarose gels and visualized under UV illumination by staining with GelRed.
A sample was regarded as methylation negative when a PCR product was obtained with the U primer set and methylation-positive samples were considered as those positive for a PCR product with the M primer set or both the M and U set. Reactions prepared with water instead of template DNA were used as a negative control.
Statistical analysis
Statistical analyses were performed with the SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA). Continuous variables were expressed as a median (centile 25; centile 75). The Kolmogorov-Smirnov test was used to examine whether the data were derived from a normally distributed population. Differences in variables were analyzed by the Kruskal-Wallis or Mann-Whitney U test. The Chi-squared test was used to compare methylation frequency of the ESR1 promoter between different groups. Univariate Cox proportional hazards regression analysis was used to determine the factors affecting survival time. All possible predictive variables were included in a multivariate Cox model with a backward stepwise method to remove the least predictive variable at each step. Survival curves were drawn using the Kaplan-Meier method and the statistical significance was determined using the log-rank test. The diagnostic value of various clinical characteristics and associated criteria was assessed by receiver operating characteristic (ROC) curves. A two-tailed value of p < 0.05 was considered as statistically significant.
Results
Basic characteristics of the training cohort
The characteristics of the enrolled participants in the training cohort are shown in table 1. There was no significant difference between HBeAg (p = 0.877), HBeAb (p = 0.881) and HBV DNA load (p = 0.769) between the ACHBLF and CHB groups. There were no significant differences among ACHBLF, CHB and HCs with regard to age (p = 0.949), sex (p = 0.924) and Cr (p = 0.281). However, significant differences were found with respect to ALT, AST, TBil, ALB, PTA and INR (p < 0.001, respectively) within the three groups. The incidence of liver cirrhosis, ascites, hepatic encephalopathy (HE) and spontaneous bacterial peritonitis in the ACHBLF group was 55.75%, 66.37%, 30.97% and 27.43%, respectively, and the mortality rate was 67.26%.
The clinical characteristics on admission to hospital were compared between survivors and non-survivors in the training cohort. The patients who died had a higher bilirubinemia level (364.70 μmol/l vs 231.90 μmol/l, p = 0.003) and MELD score (23.41 vs 19.58, p < 0.001) and also had an increased incidence of HE (40.79% vs 10.81%, p = 0.001). These data are presented in supplementary table 1.
Elevated methylation frequency of the ESR1 promoter in peripheral blood mononuclear cells of ACHBLF patients in the training cohort
ESR1 is partly associated with the pathogenesis of ACHBLF, thus, the methylation frequency of the ESR1 promoter in PBMCs of patients with ACHBLF, CHB, and HCs was assessed using the MSP technology.
Representative examples of MSP assay results are presented in figures 2A and B. A significant increase of the methylation frequency of ESR1 was found in PBMCs of patients with ACHBLF (66.37%) compared to those with CHB (21.92%, p < 0.001) and NC (11.54%, p < 0.001) (Fig. 2C). However, no differences with regard to DNA methylation frequency were found between NC and patients with CHB (p = 0.229) (Fig. 2C). The ESR1 promoter methylation frequency in PBMCs was analyzed between non-survivors and survivors of ACHBLF in the training cohort. Of the 113 patients with ACHBLF, 37 were categorized as survivors and 76 as non-survivors. The non-survivors had a significantly higher methylation frequency than survivors (77.63% vs 29.73%, p < 0.001) (Fig. 2D). Promoter hypermethylation is an epigenetic mechanism of gene silencing or decreased expression. The mRNA levels were compared between patients with methylated and unmethylated ESR1; the ESR1 mRNA levels were significantly lower in methylated cases (3.86 × 10-4 ± 3.61 × 10-4) than in unmethylated cases (8.42 × 10-4 ± 6.64 × 10-4, p < 0.001) (Fig. 2E).
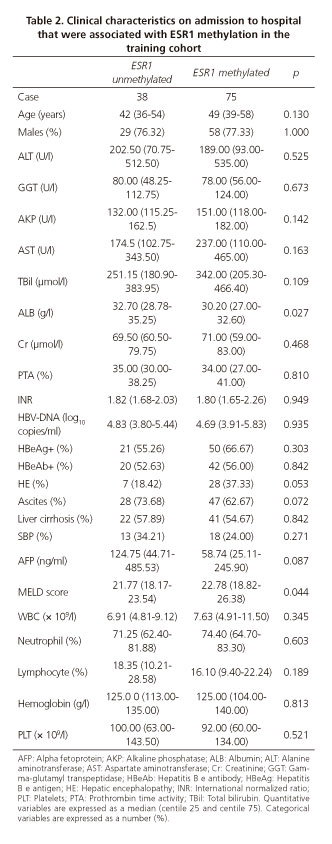
To determine whether higher ESR1 promoter methylation frequency contributed to the severity of ACHBLF, the correlations between ESR1 gene promoter methylation and clinical characteristics was analyzed in the 113 patients with ACHBLF in the training cohort. ESR1 promoter methylation was significantly associated with ALB (p = 0.027) and MELD scores (p = 0.044). However, there were no significant correlations with age, sex, HBV-DNA load, ALT, AST, alkaline phosphatase, gamma-glutamyl transpeptidase, TBil, Cr, PTA, INR, HBeAg, HBeAb, alpha fetoprotein, white blood cell, neutrophil, lymphocyte, hemoglobin and platelet levels. Moreover, no significant association was found between ESR1 promoter methylation and liver cirrhosis, ascites, HE and spontaneous bacterial peritonitis. These data suggest that elevated ESR1 promoter methylation frequency may be involved in hepatic synthetic impairment and closely associated with disease severity of ACHBLF. These data are shown in table 2.
Follow-up of ESR1 promoter methylation in patients initially diagnosed with pre-ACHBLF that progressed to ACHBLF
Eighteen patients were included in a retrospective study from the pre-ACHBLF group that developed into the ACHBLF group between September 2009 and May 2015. ACHBLF is a progressive disease (32), therefore, we evaluated the methylation status of the ESR1 promoter on the day of pre-ACHBLF diagnosis as well as on the 1st, 3rd and 7th day of ACHBLF and the day before death or the 28th day of the study. Figure 2F shows that ESR1 promoter methylation status changed during the disease course. At the condition of pre-ACHBLF, two in 15 non-survivors exhibited ESR1 promoter methylation, while the ratio reached 5:15 on the first day of ACHBLF. On the third day, two patients died and the ratio was up to 7:13, then 6:6 on the seventh day (nine patients had already died). Among the survivors group, only one patient showed ESR1 promoter methylation on the third day of ACHBLF, and this patient showed unmethylation on the seventh day and 28th day of ACHBLF; one patient in the survivors group showed ESR1 promoter methylation on the 3rd day, 7th day, and 28th day. Unfortunately, this patient died of multiple organ dysfunction syndrome on the 31st day (after three days of follow-up). These results indicate that ESR1 promoter methylation was positively correlated with the patient's severe condition.
Univariate and multivariate analysis of risk factors for 28-day mortality of ACHBLF patients in training cohort
Cox proportional hazards regression analysis was performed to identify which clinical parameters affected survival time. As shown in table 3, age (p = 0.040), TBil (p = 0.002), ALB (p = 0.048), Cr (p < 0.001), PTA (p = 0.001), INR (p < 0.001), MELD scores (p < 0.001), HE (p = 0.001) and ESR1 methylation (p < 0.001) were associated with mortality in a univariate analysis. On multivariate analysis, the only variables that were associated with 28-day mortality were MELD scores (p < 0.001), HE (p = 0.017) and ESR1 methylation (p < 0.001). Thus, indicating that HE, MELD score and ESR1 promoter methylation are independent risk factors associated with 28-day mortality in ACHBLF.
ESR1 promoter methylation as a predictor of poor prognosis of patients with ACHBLF in the training and validation cohort
Among the 113 patients with ACHBLF in the training cohort, 76 died during the 28-day follow-up. The mortality rate was 67.26% and the mean survival time was 17.18 days (standard error [SE] 0.89, 95% confidence interval [CI] 15.44-18.91). The mean survival time of patients with ACHBLF and no ESR1 promoter methylation was 25.37 days (SE 0.96, 95% CI 23.49-27.24), which was significantly higher than in patients with ESR1 promoter methylation (mean survival time 13.03 days, SE 0.93, 95% CI 11.21-14.85, p < 0.001) (Fig. 3). ESR1 promoter methylation had a sensitivity of 85.53%, specificity of 72.97%, positive likelihood ratio (LR+) of 3.16 and negative likelihood ratio (LR-) of 0.20 when used to predict 28-day mortality in ACHBLF. The area under the curve (AUC) was 0.79 (SE 0.04, 95% CI 0.71-0.86, p < 0.001) (Fig. 3B). In order to validate the accuracy of ESR1 promoter methylation as a predictive biomarker for poor prognosis of patients with ACHBLF, another 37 patients with CHBLF were enrolled as a validation cohort. The basic characteristics of the patients are shown in supplementary table 2. Figure 3C shows that ACHBLF patients with ESR1 promoter methylation had a significantly shorter survival time (13.83 days, SE 1.56, 95% CI 10.76-16.89) than patients without ESR1 promoter methylation (22.25 days, SE 3.52, 95% CI 15.34-29.16, p = 0.015). This data was similar to that seen in the training cohort. The AUC of ESR1 promoter methylation was 0.73 (SE 0.08, 95% CI 0.56-0.87, p = 0.005) (Fig. 3D). Therefore, these results demonstrated that ESR1 promoter methylation could serve as a promising biomarker for 28-day mortality in patients with ACHBLF.
Discussion
ACHBLF is a medical ailment and the associated short and medium-term mortality is between 50% and 90% (1). Several prognostic models have been proposed but none of these models accurately predict the mortality in ACHBLF (31,33). In addition, some logistic regression models are based on 3-month mortality (34,35), which does not reflect the severity of ACHBLF and the urgent time frame for liver transplantation. The MELD score is the most commonly used prediction model and has been used for patients with ACHBLF (36). However, the MELD score was originally established to evaluate short-term mortality risk in patients with cirrhosis that required a transjugular intrahepatic portosystemic shunt to be fitted (31). The use of this method for the prediction of prognosis in ACHBLF is controversial (37,38). Therefore, biomarkers with a higher sensitivity and specificity are needed to predict ACHBLF prognosis at an early stage in order to offer the best opportunity for liver transplantation.
In this study, we report a significant increase of ESR1 promoter methylation frequency in PBMCs from patients with ACHBLF compared with those with CHB and HCs. Lower ALB and a higher MELD score were significantly associated with ESR1 promoter methylation. Moreover, ESR1 promoter methylation frequency increased with ACHBLF progression. These results strongly indicated that ESR1 methylation contributed to hepatic synthetic impairment and was closely associated with ACHBLF disease severity. Moreover, ESR1 promoter methylation was one of the independent factors to predict 28-day mortality. In addition, ESR1 promoter methylation had a significant power to predict 28-day mortality from ACHBLF. These data demonstrate that ESR1 promoter methylation may be involved in the pathogenesis of ACHBLF and serve as a predictive biomarker of prognosis in patients with ACHBLF.
ESR1 interacts with the hepatocyte nuclear factor-4α to HBV enhancer I to suppress HBV gene transcription (39). In fact, the level of ESR1 in the cytosol of PBMCs is significantly lower in asymptomatic HBV carriers and patients with CHB than in healthy controls (40). HBV reactivation is still the leading cause of ACLF in the East. HBV-infected hepatocytes often exhibit an abnormal epigenetic status (41). Therefore, we hypothesized that ESR1 promoter methylation may be involved in the pathophysiological mechanism of ACHBLF. There are five aspects that support this notion. First, the frequency of ESR1 promoter methylation in PBMCs from patients with ACHBLF was significantly higher than that in patients with CHB and HCs. Second, ESR1 methylation frequency was correlated with ALB and the MELD score and increased with ACHBLF progression. Third, ESR1 promoter methylation was one of the independent predictive factors for 28-day mortality. Fourth, the frequency of ESR1 promoter methylation in non-survivors was significantly higher than that of survivors. Lastly, ESR1 promoter methylation had a significant power to predict 28-day mortality from ACHBLF both in the training and the validation cohort. Increased ESR1 promoter methylation could play a crucial role in ACHBLF pathogenesis. However, there was no significant correlation between ESR1 promoter methylation and HBV DNA load. Therefore, the exact mechanism of ESR1 methylation in ACHBLF should be extensively studied in the future.
This study is the first to show that aberrant ESR1 promoter methylation might serve as a prognostic biomarker for ACHBLF. The mean survival time of patients with ACHBLF with ESR1 promoter methylation in this study was much shorter than in those without ESR1 promoter methylation. In China, the 28-day mortality rate from ACHBLF is high and there are limited liver donors. Therefore, ESR1 promoter methylation studies could help to identify those patients that are most in need of a liver transplantation.
This study has several limitations. The relatively small number of survivors may affect the accuracy of the conclusions due to the real life situation in China with regard to ACHBLF epidemiology data. In addition, the Qilu hospital is a tertiary care hospital in the Shandong province, thus patients referred to our hospital have a more serious condition. Our conclusion should be authenticated in a multicenter, large-scale cohort. In addition, our data were generated from PBMCs which may not be representative of the intrahepatic condition. However, we were unable to confirm our conclusion using liver biopsies. ACHBLF is a severe life-threatening clinical syndrome with a high mortality rate between 50% and 90%, especially with more serious coagulation disorders. We were unable to encourage patients to undergo a liver biopsy and none of the patients underwent liver transplantation. Therefore, there was no liver tissue available from these patients. Our conclusions could be confirmed in a future study using tissue from patients who undergo a liver transplantation.
In this study, the MSP method was used to analyze the methylation status of the ESR1 gene promoter due to the availability of the MSP method in clinical laboratories and the fact that the results are obtained on the same day. Although the sensitivity and specificity of MSP is not satisfactory, it can be used to select the patients with a methylation status requiring further examination. However, a quantitative method could provide more accurate information about the methylation pattern, and this method should be performed in future studies.
In summary, our study was the first to reveal that abnormal ESR1 methylation occurred in patients with ACHBLF. ESR1 methylation might potentially serve as a prognostic biomarker for ACHBLF. This study provides a new direction for further evaluation and treatment of ACHBLF.
References
1. Sarin SK, Kedarisetty CK, Abbas Z, et al. Acute-on-chronic liver failure: Consensus recommendations of the Asian Pacific Association for the Study of the Liver (APASL) 2014. Hepatol Int 2014;8:453-71. DOI: 10.1007/s12072-014-9580-2. [ Links ]
2. Xu L, Tu Z, Xu G, et al. Epirubicin directly promotes hepatitis B virus (HBV) replication in stable HBV-expressing cell lines: A novel mechanism of HBV reactivation following anticancer chemotherapy. Mol Med Rep 2014;9:1345-50. DOI: 10.3892/mmr.2014.1973. [ Links ]
3. Mikulska M, Nicolini L, Signori A, et al. Hepatitis B reactivation in HBsAg-negative/HBcAb-positive allogeneic haematopoietic stem cell transplant recipients: Risk factors and outcome. Clin Microbiol Infect 2014;20:O694-701. DOI: 10.1111/1469-0691.12611. [ Links ]
4. Finkenstedt A, Nachbaur K, Zoller H, et al. Acute-on-chronic liver failure: Excellent outcomes after liver transplantation but high mortality on the wait list. Liver Transpl 2013;19:879-86. DOI: 10.1002/lt.23678. [ Links ]
5. Zheng YB, Huang ZL, Wu ZB, et al. Dynamic changes of clinical features that predict the prognosis of acute-on-chronic hepatitis B liver failure: A retrospective cohort study. Int J Med Sci 2013;10:1658-64. DOI: 10.7150/ijms.6415. [ Links ]
6. Kamath PS, Kim WR. The model for end-stage liver disease (MELD). Hepatology 2007;45:797-805. DOI: 10.1002/hep.21563. [ Links ]
7. Shimizu I, Kohno N, Tamaki K, et al. Female hepatology: Favorable role of estrogen in chronic liver disease with hepatitis B virus infection. World J Gastroenterol 2007;13:4295-305. DOI: 10.3748/wjg.v13.i32.4295. [ Links ]
8. Hishida M, Nomoto S, Inokawa Y, et al. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol 2013;43:88-94. DOI: 10.3892/ijo.2013.1951. [ Links ]
9. Ahlbory-Dieker DL, Stride BD, Leder G, et al. DNA binding by estrogen receptor-alpha is essential for the transcriptional response to estrogen in the liver and the uterus. Mol Endocrinol 2009;23:1544-55. DOI: 10.1210/me.2009-0045. [ Links ]
10. Giannitrapani L, Soresi M, La Spada E, et al. Sex hormones and risk of liver tumor. Ann NY Acad Sci 2006;1089:228-36. DOI: 10.1196/annals.1386.044. [ Links ]
11. Villa E, Baldini GM, Rossini GP, et al. Ethanol-induced increase in cytosolic estrogen receptors in human male liver: A possible explanation for biochemical feminization in chronic liver disease due to alcohol. Hepatology 1988;8:1610-4. DOI: 10.1002/hep.1840080623. [ Links ]
12. Porter LE EM, Van Thiel DH, Eagon PK. Hepatic estrogen receptor in human liver disease. Gastroenterology 1987;92:735-45. DOI: 10.1016/0016-5085(87)90026-6. [ Links ]
13. Nagasue N IA, Yukaya H, Ogawa Y. Estrogen receptors in hepatocellular carcinoma. Cancer 1986;57:87-91. DOI:10.1002/1097-0142(19860101)57:1<87::AID-CNCR2820570118> 3.0.CO;2-K. [ Links ]
14. Villa E, Dugani A, Moles A, et al. Variant liver estrogen receptor transcripts already occur at an early stage of chronic liver disease. Hepatology 1998;27:983-8. DOI: 10.1002/hep.510270413. [ Links ]
15. Wang SH, Chen PJ, Yeh SH. Gender disparity in chronic hepatitis B: Mechanisms of sex hormones. J Gastroenterol Hepatol 2015;30:1237-45. DOI: 10.1111/jgh.12934. [ Links ]
16. Dou CY, Fan YC, Cao CJ, et al. Sera DNA methylation of CDH1, DNMT3b and ESR1 promoters as biomarker for the early diagnosis of hepatitis B virus-related hepatocellular carcinoma. Dig Dis Sci 2016;61:1130-8. DOI: 10.1007/s10620-015-3975-3. [ Links ]
17. Perret C. Methylation profile as a new tool for classification of hepatocellular carcinoma. J Hepatol 2011;54:602-3. DOI: 10.1016/j.jhep.2010.10.015. [ Links ]
18. Gao S, Ji XF, Li F, et al. Aberrant DNA methylation of G-protein-coupled bile acid receptor Gpbar1 predicts prognosis of acute-on-chronic hepatitis B liver failure. J Viral Hepatitis 2015;22:112-9. DOI: 10.1111/jvh.12277. [ Links ]
19. Archer KJ, Mas VR, Maluf DG, et al. High-throughput assessment of CpG site methylation for distinguishing between HCV-cirrhosis and HCV-associated hepatocellular carcinoma. Mol Genet Genomics 2010;283:341-9. DOI: 10.1007/s00438-010-0522-y. [ Links ]
20. Dai B, Geng L, Yu Y, et al. Methylation patterns of estrogen receptor alpha promoter correlate with estrogen receptor alpha expression and clinicopathological factors in hepatocellular carcinoma. Exp Biol Med (Maywood) 2014;239:883-90. DOI: 10.1177/1535370214536651. [ Links ]
21. Khakpour G, Pooladi A, Izadi P, et al. DNA methylation as a promising landscape: A simple blood test for breast cancer prediction. Tumour Biol 2015;36:4905-12. DOI: 10.1007/s13277-015-3567-z. [ Links ]
22. Joensuu EI, Abdel-Rahman WM, Ollikainen M, et al. Epigenetic signatures of familial cancer are characteristic of tumor type and family category. Cancer Res 2008;68:4597-605. DOI: 10.1158/0008-5472.CAN-07-6645. [ Links ]
23. Zheng DL, Zhang L, Cheng N, et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol 2009;50:377-87. DOI: 10.1016/j.jhep.2008.10.019. [ Links ]
24. Ameer F, Munir R, Usman H, et al. Lipid-load in peripheral blood mononuclear cells: Impact of food-consumption, dietary-macronutrients, extracellular lipid availability and demographic factors. Biochimie 2017;135:104-10. DOI: 10.1016/j.biochi.2017.01.015. [ Links ]
25. Hotamisligil GS. Inflammation and metabolic disorders. Nature 2006;444:860-7. DOI: 10.1038/nature05485. [ Links ]
26. Bouwens M, Afman LA, Müller M. Fasting induces changes in peripheral blood mononuclear cell gene expression profiles related to increases in fatty acid beta-oxidation: Functional role of peroxisome proliferator activated receptor alpha in human peripheral blood mononuclear cells. Am J Clin Nutr 2007;86:1515-23. [ Links ]
27. Sarin SK, Kumar A, Almeida JA, et al. Acute-on-chronic liver failure: Consensus recommendations of the Asian Pacific Association for the study of the liver (APASL). Hepatol Int 2009;3:269-82. DOI: 10.1007/s12072-008-9106-x. [ Links ]
28. Xia Q, Dai X, Zhang Y, et al. A modified MELD model for Chinese pre-ACLF and ACLF patients and it reveals poor prognosis in pre-ACLF patients. PLoS One 2013;8:e64379. DOI: 10.1371/journal.pone.0064379. [ Links ]
29. Lok AS, McMahon BJ. Chronic hepatitis B: Update 2009. Hepatology 2009;50:661-2. DOI: 10.1002/hep.23190. [ Links ]
30. Shrestha SM. Liver cirrhosis and hepatocellular carcinoma in hepatic vena cava disease, a liver disease caused by obstruction of inferior vena cava. Hepatol Int 2009;3:392-402. DOI: 10.1007/s12072-009-9122-5. [ Links ]
31. Malinchoc M, Kamath PS, Gordon FD, et al. A model to predict poor survival in patients undergoing transjugular intrahepatic portosystemic shunts. Hepatology 2000;31:864-71. DOI: 10.1053/he.2000.5852. [ Links ]
32. Ampuero J. Acute-on-chronic liver failure: A time to step forward. Rev Esp Enferm Dig 2017;109:397-8. DOI: 10.17235/reed.2017.5054/2017. [ Links ]
33. Xun YH, Shi JP, Li CQ, et al. Prognostic performance of a series of model for end-stage liver disease and respective Delta scores in patients with hepatitis B acute-on-chronic liver failure. Mol Med Rep 2014;9:1559-68. [ Links ]
34. Sun QF, Ding JG, Xu DZ, et al. Prediction of the prognosis of patients with acute-on-chronic hepatitis B liver failure using the model for end-stage liver disease scoring system and a novel logistic regression model. J Viral Hepat 2009;16:464-70. DOI: 10.1111/j.1365-2893.2008.01046.x. [ Links ]
35. Zheng MH, Shi KQ, Fan YC, et al. A model to determine 3-month mortality risk in patients with acute-on-chronic hepatitis B liver gailure. Clin Gastroenterol Hepatol 2011;9:351-6.e3. DOI: 10.1016/j.cgh.2010.12.027. [ Links ]
36. Lai J, Lin CS, Yang L, et al. Pretreatment HBsAg level and an early decrease in MELD score predict prognosis to lamivudine treatment for HBeAg-negative acute-on-chronic liver failure. Clin Res Hepatol Gastroenterol 2014;38:331-6. DOI: 10.1016/j.clinre.2013.10.012. [ Links ]
37. Barosa R, Roque Ramos L, Patita M, et al. CLIF-C ACLF score is a better mortality predictor than MELD, MELD-Na and CTP in patients with Acute on chronic liver failure admitted to the ward. Rev Esp Enferm Dig 2017;109:399-405. DOI: 10.17235/reed.2017.4701/2016. [ Links ]
38. Wang SH, Yeh SH, Lin WH, et al. Estrogen receptor alpha represses transcription of HBV genes via interaction with hepatocyte nuclear factor 4alpha. Gastroenterology 2012;142:989-98e4. DOI: 10.1053/j.gastro.2011.12.045. [ Links ]
39. Mizoguchi Y, Takeda H, Sakagami Y, et al. Estradiol receptors in the cytosol of peripheral blood mononuclear cells in hepatitis B virus carriers treated with interferon-alpha. Gastroenterol Jpn 1989;24:373-9. [ Links ]
40. Zhu R, Zhang JS, Zhu YZ, et al. HBx-induced androgen receptor expression in HBV-associated hepatocarcinoma is independent of the methylation status of its promoter. Histol Histopathol 2011;26:23-35. [ Links ]
![]() Correspondence:
Correspondence:
Kai Wang.
Department of Hepatology.
Qilu Hospital of Shandong University and Hepatology Institute of Shandong University.
Wenhuaxi Road 107#. 250012 Jinan, China
e-mail: wangdoc876@126.com;
wangdoc2010@163.com
Received: 12-05-2016
Accepted: 25-07-2017