Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
International Microbiology
versión impresa ISSN 1139-6709
INT. MICROBIOL. vol.7 no.1 mar. 2004
| RESEARCH REVIEW | ||||
|
| ||||
| IS200: an old and still bacterial transposon | |||
| Summary. IS200 is a mobile element found in a variety of eubacterial genera, such as Salmonella, Escherichia, Shigella, Vibrio, Enterococcus, Clostridium, Helicobacter, and Actinobacillus. In addition, IS200-like elements are found in archaea. IS200 elements are very small (707-711 bp) and contain a single gene. Cladograms constructed with IS200 DNA sequences suggest that IS200 has not spread among eubacteria by horizontal transfer; thus it may be an ancestral component of the bacterial genome. Self-restraint may have favored this evolutionary endurance; in fact, unlike typical mobile elements, IS200 transposes rarely. Tight repression of transposase synthesis is achieved by a combination of mechanisms: inefficient transcription, protection from impinging transcription by a transcriptional terminator, and repression of translation by a stem-loop mRNA structure. A consequence of IS200 self-restraint is that the number and distribution of IS200 elements remain fairly constant in natural populations of bacteria. This stability makes IS200 a suitable molecular marker for epidemiological and ecological studies, especially when the number of IS200 copies is high. In Salmonella enterica, IS200 fingerprinting is extensively used for strain discrimination. [Int Microbiol 2004; 7(1):3-12] Key words: transposition · DNA rearrangements · genome evolution · parasite attenuation · IS200 fingerprints | ||||
| IS200: un antiguo y tranquilo transposón bacteriano Palabras clave: transposición · reordenamientos del DNA · evolución del genoma · atenuación de parásitos · perfiles IS200 | IS200: um antigo e calmo transposon bacteriano Palavras chave: transposição · reordenamentos de DNA · evolução do genoma · atenuação de parasitos · perfis IS200 |
Introduction
Transposons are mobile DNA sequences able to insert themselves at new locations in the genome. Unlike homologous recombination, transposition does not require homology between the transposon and its target site. Aside from transposition, mobile DNA elements can promote other genome rearrangements, such as duplications, deletions, and inversions [46,53]. Because of their replication at the genome's expense and their mutational capability, transposons can be viewed as parasite DNAs. However, at the population level, they can serve as sources of genetic variation, thereby conveying adaptive value. Given this dual nature, the evolutionary success of a given transposon requires a delicate equilibrium: its contribution to population polymorphism must be high but its impact on individual fitness must be low. Bacterial transposons thatare not known to carry genes other than those required for transposition are usually known as "insertion sequences." Such elements are common in bacterial chromosomes, and are also found in plasmids and phage genomes [15,17,22,39]. IS200 is an insertion sequence originally found in Salmonella [34], and later described in other bacterial genera. The element is small, devoid of the symmetric ends typical of many bacterial ISs, and reluctant to transpose. While IS200 might initially be viewed as an inert, dull example of a transposon, its distribution among bacterial genera suggests that the element has a long evolutionary history. Because attenuation is a key factor in the long-term survival of parasites [42], the evolutionary endurance of IS200 might have been favored by its low activity. IS200 may provide an example of the evolutionary advantage of parasite self-restraint. Below, we summarize two decades of research on IS200, with emphasis on its many unsolved questions and paradoxes.
Discovery of IS200
Among the hundreds of mutations characterized in the Salmonella histidine operon between 1960 and 1970, hisD984 was a peculiar case. The mutation had been isolated by diethylsulfate mutagenesis [38], a procedure that usually yields base substitutions. Not surprisingly, hisD984 mapped as a point mutation [26]. However, the mutation did not revert, neither spontaneously nor upon treatment with mutagens [26]. Equally puzzling was the fact that, unlike nonsense codons and frameshifts, hisD984 was completely polar on the downstream gene hisC [28]. These contradictory features were finally understood when Steve Lam, a graduate student in John Roth's laboratory at the University of Utah, discovered that hisD984 was an insertion mutation [34]. Lam analyzed the hisD984 mutation by Southern hybridization, using the only probe available at the time, an M13 derivative carrying hisG, hisD and hisC of Salmonella [1]. An extra piece of DNA, some 700-bp long, was found inserted in hisD [34]. Hybridization studies also showed that additional copies of this DNA piece were present in the Salmonella genome, suggesting that hisD984 was a typical insertion mutation, caused by a mobile DNA element [34]. The absence of cross-hybridization with Escherichia coli DNA suggested that the transposable element involved should be novel; it was registered as such in the IS database and given the number 200 [34]. The same study investigated the presence of IS200 in Salmonella isolates of diverse origin, and found that the element was present in most of them. In contrast, IS200 was not found in DNA preparations from Escherichia, Citrobacter, Proteus, Klebsiella, and Serratia. Only Shigella DNA hybridized weakly with the IS200 probe. This highly biased distribution suggested that IS200 was a Salmonella-specific insertion element [34]. Although further surveys, carried out with larger and more diverse strain samples, have indicated that this bias is not absolute [8,23], the skewed distribution of IS200 in enteric bacteria remains an enigmatic observation and will be discussed below.
Structure of IS200
The fact that the DNA ends sequenced by Lam and Roth [36] had a slight degree of homology with IS630, an IS found in Shigella, prompted the hypothesis that IS200 was a deletion derivative of IS630 [41]. The idea that IS200 was a distinct element was not fully accepted until the hisD984::IS200 insertion was thoroughly sequenced-albeit with a frameshift-in 1991 [24]. Soon after, sequencing of several independent IS200 elements permitted a reliable description of the IS200 structure [3]. Because of the reluctance of IS200 to transpose, we wondered whether some copies of the element-or perhaps even many-might be defective. Hence, both genomic copies and hops were sequenced in order to increase the chances that a wild-type element could be found among the elements sampled. The actual result, however, was that IS200 hops and IS200 elements residing in the Salmonella genome were all similar or identical, thereby ruling out the accumulation of defective copies [3].
IS200 is one of the smallest transposable elements found in bacteria, and shows a highly compact structure, with little spare room. Different copies have slightly different lengths, from 707 to 711 bp, and all lack terminal repeats, direct or inverse [3]. This trait is rare among bacterial insertion sequences, but it is not exclusive of IS200: terminal repeats are also absent from elements like IS91 and IS110 [39]. Sequence analysis also indicated that the ~50 bp located at each end of IS200 are highly conserved [3]. Other conserved regions are the single IS200 gene, which encodes a protein of 16 kDa [3,8], and two structures that play a role in IS200 gene expression: a RNA stem-loop that occludes the ribosome-binding-site of the IS200 single gene, and a Rho-independent transcription terminator, which, surprisingly, appears to overlap the IS200 promoter (Fig. 1) [3,4].
Two comprehensive reviews on insertion elements, published by Jacques Mahillon and Michael Chandler in 1998 and 2002, respectively, classified IS200 and the Helicobacter element IS605 in a single group or family [15,39]. Unlike IS200, IS605 contains two open reading frames, of which only one is related to IS200 [15,39]. Two other elements, IS1253A from Dichelobacter nodosus, and ISH1-8 from halobacteria, are also included in the IS200/IS605 family [15,39].
Search for IS200 hops
During the 1970s, work in E. coli had established the paradigm that mobile DNA elements cause a substantial proportion of spontaneous mutations in bacteria. For instance, a review contained the following statement: "If selection for inactivation of a gene is made, many of the mutants will probably be ‘insertion mutants'" [17]. Based on this paradigm, experiments searching for the presence of IS200 insertions in the his operon of Salmonella were carried out. Taking advantage of the osmosensitivity of histidine-constitutive strains [52], isolates able to grow on high-salt media were selected, and His¯mutants were sought among them. The His¯ auxotrophs obtained were then characterized by reversion analysis, complementation, and mapping. Any point mutation with strong or absolute polarity would qualify as a potential IS200 hop. The screen seemed appropriate, because the existence of the mutation hisD984 indicated that the his operon contained at least one IS200 target. Furthermore, the selection strategy favored the detection of isolates carrying polar blocks upstream of the osmosensitivity-causing genes hisH and hisF, and the mutation hisD984::IS200 was strongly polar. However, the search for IS200 hops failed: genetic analysis of over 2,500 his polar mutations revealed only deletions, frameshifts, and nonsense codons, and a few bizarre mutations that did not fit any of these types were discarded by physical analysis [14].
At that stage, the failure to detect IS200 hops among histidine auxotrophs was a remarkable observation, but could be still considered inconclusive. For instance, one might wonder whether the selection employed could somehow hamper IS200 hopping. However, later screens and selections of various types failed also (C.R. Beuzón, Ph.D. Thesis, University of Seville, Spain, 1996). Some of these searches were based on the use of plasmids devised to serve as "IS traps", and had proven useful in E. coli and other bacteria [51,58]. Others, like the PCR assay summarized in Fig. 2, should have detected IS200 hopping to hisD even if the transposition frequency was extremely low. All trials failed, providing evidence that, unlike other insertion elements, IS200 does not transpose at detectable frequencies (C. R. Beuzón, Ph.D. Thesis, 1996). In addition, genetic trials supported the conclusion that the genome of Salmonella does not contain other active transposons. This view is illustrated by the following anecdote: in one experiment, an element was found inserted in the transposon-trapping plasmid pRAB2 [51]. Not surprisingly, another element was also found on the chromosome. When the pedigree of the strain was traced back, it turned out that one if its ancestors had hosted an F', which was probably the original source of the element (C. R. Beuzón and J. Casadesús, unpublished data).
Ken Haack, a graduate student in John Roth's laboratory during the early 1990s, sought IS200-induced mutants using a different approach. Haack constructed composite transposons that carried a kanamycin-resistance determinant flanked by IS200 elements in direct or inverse orientation. Plasmids carrying these IS200-flanked transposons were transduced to polA hosts (to prevent plasmid maintenance), and Kmr transductants were selected. Transductants lacking the plasmid-borne Apr determinant were subjected to physical analysis. To Haack's surprise, none of the Kmr transductants examined carried a "simple" insertion of an IS200-flanked transposon (K.R. Haack, Ph.D. Thesis, University of Utah, Salt Lake City, Utah, 1995). This failure sharply contrasts with the ability of composite mobile elements-naturally existing, such as Tn9 [25], or artificially made, such as Tn2700 [60]-to transpose, and provided further evidence that IS200 does not undergo detectable frequencies of transposition. To date, insertions of IS200 have been only found, some of them accidentally, in the chromosomal genes hisD [34] and gpt [47], and in the plasmid-borne pef operon [3].
The low activity of IS200 is observed not only under laboratory conditions. Surveys carried out in collaboration with Salvatore Rubino's group at the University of Sassari led us to the conclusion that natural populations of Salmonella do not undergo high rates of IS200 hopping. In those experiments, isolates of Salmonella abortusovis were collected from sheep in several towns of the island of Sardinia. The numbers and locations of their IS200 elements were compared with those of isolates of S. abortusovis obtained from Sardinian flocks 30 years earlier. Strains with identical IS200 profiles were found in the two collections, confirming that IS200 transposes rarely in the wild [56]. Is infrequent transposition truly surprising? Or is it that our view of the transposon world is biased towards its most active members? An insertion element rarely transposes if the genome of its host is scarce in targets, as shown in Salmonella for IS30, a common element in E. coli [12]. Easily identifiable mutations (e.g. auxotrophies) can be also rare if the element avoids transcribed regions, a behavior displayed by Mu [19], Tn10 [13] and Tn7 [20]. Furthermore, as relatively simple genetic entities, insertion elements often require host functions to perform transposition [22,39]. Thus, variations in the availability of specific cell functions (e.g., those ancillary for transposition) can affect transposon activity and influence the fate of the element in a new host. Finally, elements with intrinsically low activity can be expected to exist; in fact, self-restraint can be viewed as the safest strategy for transposon endurance [21]. Thus transposable elements with high frequencies of transposition may be the exception rather than the rule, and IS200 may be just an ordinary transposon with extremely low activity.
A caveat to the above conclusion is the observation, first made by Steve Lam and John Roth, that IS200 hops are detected in stab cultures [34]. An additional finding, tentatively documented with simple statistical analysis, is that if a hop is detected in a stab, additional insertions will probably be found (C.R. Beuzón, Ph.D. Thesis, 1996). An appealing possibility is that IS200 undergoes stochastic activation: in a given cell, a small difference in IS200 mRNA content might be amplified, causing a burst of transposase synthesis.
However, bursts of transposition are not observed in batch cultures, thereby raising the possibility that IS200 activation is not random, and that growth conditions unique to stab cultures might trigger transposase synthesis. Whatever the case, environmental influences, such as anaerobiosis, stationary phase, high or low osmolarity, heat shock, and cell density, do not seem to play any role in the process (K.R. Haack and J. Casadesús, unpublished). The same conclusion applies to host functions, including DNA adenine methylation, DNA gyrase, and nucleoid proteins HU and IHF (D. Chessa, C. R. Beuzón and J. Casadesús, unpublished).
DNA rearrangements induced by IS200
When Haack and Roth analyzed Kmr isolates generated by transfer of IS200-flanked transposons from a plasmid to the Salmonella chromosome, they found several kinds of DNA rearrangements (K.R. Haack, Ph.D. Thesis, 1995). The closest to a hop was a complex insertion in the leu operon, whose structure could be tentatively explained by replicative transposition, followed by additional DNA rearrangements (K.R. Haack, Ph.D. Thesis, 1995). Complex insertions of this kind were only generated by the inverse order construct. In contrast, the transposon flanked by direct repeats of IS200 mostly yielded recombinants in which the plasmid-borne transposon had recombined with a chromosomal IS200 copy. Although this type of event was RecA-dependent, Haack and Roth observed that it could be stimulated by overproduction of IS200 transposase (K.R. Haack, Ph.D. Thesis, 1995). Transposase-stimulated recombination was also detected between chromosomal IS200 copies, suggesting that DNA nicks made by the IS200 transposase at the ends of IS200 elements might stimulate homologous recombination [27].
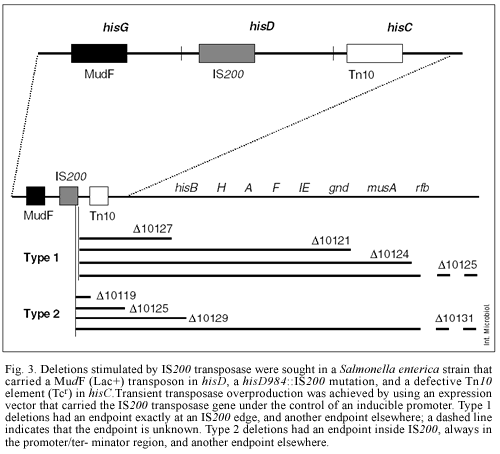
Another type of chromosome rearrangement is deletion formation (C.R. Beuzón, Ph.D. Thesis, 1996). During our searches for IS200 hops in stab cultures, we observed band shifts among IS200-hybridizing fragments, and obtained evidence that some of the shifts were caused by chromosomal deletions near an IS200 element. The possibility that IS200, like other transposable elements, generates deletions was thus considered. To examine this hypothesis, strains carrying antibiotic-resistance markers in hisG and hisC, as well as an IS200 element in hisD, were constructed (Fig. 3). Deletions could be detected by loss of one or both antibiotic-resistance markers, and the use of Bochner-Maloy plates permitted the positive selection of Tcs derivatives. Deletions that extended towards one side of IS200 were sought by isolating Tcs derivatives that still retained the Kmr marker. Such deletions occurred spontaneously at very low frequency (C. R. Beuzón, Ph.D. Thesis, 1996). However, Haack made the remarkable observation that deletion formation increased in the presence of a plasmid that overproduced IS200 transposase (K.R. Haack, Ph.D. Thesis, 1995). Genetic mapping of the deletions, followed by physical analysis of a subset of them, showed that the deletions were heterogeneous. One type had one endpoint exactly at the end of the element, and another endpoint elsewhere on the chromosome, sometimes at a distance of several kb (C.R. Beuzón, Ph.D. Thesis, 1996). The formation of these deletions can be tentatively explained as the result of transposase activity, as described for other transposable elements [22]. In the context of IS200 research, however, they are of special interest: the fact that their formation is promoted by overproduction of the single IS200 product provides indirect evidence that this polypeptide may be a DNA-cutting enzyme, perhaps a transposase.
The second type of deletion had an endpoint within IS200, always in the inverted repeats that embody a transcriptional terminator, and removed 10-30 bp of IS200 DNA. Because the transcriptional terminator overlaps with the IS200 promoter, these deletions can fuse the IS200 transposase gene to foreign promoters, and thus could activate IS200 transposition. The occurrence of deletions in the same stabs in which IS200 hops had been found fuels the speculation that fusion of an IS200 transposase gene to a foreign promoter might provide transposase in trans to other IS200 copies (C.R. Beuzón, Ph.D. Thesis, 1996). An alternative possibility, however, is that such deletions are by-products of transposase activity, rather than the cause of transposase overproduction.
Plasmids that overproduced the IS200 16-kDa protein were unstable and suffered DNA rearrangements at high rates. The "left", promoter-proximal end of IS200 appeared to be more prone to rearrangement than the right end. Deletions were especially common, but inversions were also detected (C.R. Beuzón and J. Casadesús, unpublished).
Target specificity of IS200
The difficulty to obtain IS200 hops makes it difficult to compare independent IS200 insertion sites, and thus to search for homologies among the targets. However, if the sequences of the few IS200 hops obtained are piled up with those of genomic copies of IS200, then the tentative conclusion is that IS200 insertion sites do not show significant homology, but all are A+T-rich. In fact, in the neighboring 10-15 bp that flank every IS200 element, 76-95% are A+T base pairs [3]. Thus, IS200 may have regional specificity for A+T-rich DNA tracts, like other insertion elements [44,63]. In the few cases in which the IS200 element sequenced is a hop (and not a genomic copy), the boundaries of the insertion can be compared to those of the wild-type gene. In such cases, a 2-bp duplication is found at the insertion site [3]. Thus IS200 may duplicate 2 bp of host DNA upon transposition.
Expression of the IS200 transposase gene
Early attempts to detect IS200-derived mRNA by either Northern hybridization or primer extension failed with no exception. The strains used were the standard wild-type S. typhimurium LT2, which contains six copies of IS200, S. typhimurium SL4510 (ten copies), a S. typhi strain with 25 or more IS200 elements [23], and an LT2 derivative carrying the hisD984::IS200 mutation on a multicopy plasmid. Since the experiments had been carried out under conditions that permitted the detection of rare RNAs, it seemed that expression of the IS200 single gene was tightly repressed.
Cloning of the single IS200 gene on an expression vector permitted the detection of IS200-derived RNA, provided that some 100 bp of the left end of IS200 had been deleted [4]. The same construction permitted SDS-PAGE detection of the single 16-kDa polypeptide encoded by IS200 [4]. The idea that this product is the IS200 transposase is based on Ken Haack's finding that its overproduction increases deletion formation (K.R. Haack, Ph.D. Thesis, 1995). More direct evidence is still awaiting, because our ignorance of the conditions that permit IS200 hopping has hampered a direct genetic test; mutations in the putative transposase gene should render a non-transposing element.
The extremely low expression of the IS200 transposase gene seems to rely on several overlapping mechanisms. One is the existence of a weak promoter, a feature common to most transposase genes [39]. In addition, IS200 transposase synthesis is kept low by a stem-loop structure formed in IS200 mRNA; this structure represses translation of the transposase gene by occluding the ribosome-binding site [4]. The stability of the IS200 stem-loop is such that the structure remains double-stranded at 60ºC in vitro [4]. The existence of strong translational repression implies that the rare transcripts initiated at the weak IS200 promoter will not be available for translation unless the mRNA stem-loop is either disrupted or cut (Fig. 4). Mechanisms for disruption or removal of RNA secondary structures have been discussed in the literature [43], and the possibility that they operate in IS200 is appealing. However, trials using a lac reporter gene fused to the IS200 ORF have so far failed to reveal any laboratory conditions in which expression is increased [4]. A caveat is that laboratory conditions may not reproduce the natural situations that induce IS200 transposition. Another limitation is that lac expression trials are not feasible in stabs, the only environment where IS200 appears to have arisen.
Another mechanism prevents the expression of the IS200 tnpA gene from foreign promoters (Fig. 4). If transcribed, two inverted repeats located upstream of the IS200 transposase gene form a transcriptional terminator [4]. This terminator, first described by Lam and Roth [36], is active in both orientations and probably belongs to the Rho-independent type [4]. If an impinging transcript enters IS200, termination at this site will prevent transcription of the transposase gene. The terminator region overlaps with the IS200 tnpA promoter, and serves as a hotspot for the formation of deletions that can fuse the IS200 transposase gene to a foreign promoter. Thus, in a fusion event of this kind, both the IS200 structure that prevents inward transcription and the native IS200 promoter will be destroyed. Such a fusion might place the IS200 transposase gene under the control of a foreign promoter, potentially leading to IS200 activation. However, this possibility remains a matter of speculation, and none of the deletions so far characterized at the histidine operon actually resulted in IS200 hopping. However, deletions that do not remove the translational repressor cannot be expected to yield high levels of transposase (see Fig. 4).
Host range, distribution and evolution of IS200 elements
When Lam and Roth first reported the existence of IS200, they found that the standard strain LT2 had six copies of the element, whereas other S. typhimurium strains had different copy numbers [34]. Most Salmonella isolates from subgenus I proved to contain IS200, but exceptions were also found [34]. The six IS200 copies of strain LT2 are all on the chromosome. Steve Lam determined their locations by using an imaginative procedure with the hallmark of John Roth's lab. A collection of 500 random Tn10 insertions was constructed, and genomic DNA preparations were made from them. The DNA preparations were then digested with PvuII, a restriction enzyme that cuts within IS200 but not within Tn10, and hybridized against an IS200 probe. Whenever a PvuII fragment contained a Tn10 element, its size increased: thus, an altered hybridization pattern was found. Genetic mapping of Tn10 insertions that had caused shifts of Southern hybridization bands could then give the approximate locations of IS200 elements [35]. Despite the limitations imposed by Tn10 insertion preferences, most of the IS200 locations obtained by Lam and Roth were remarkably precise, as indicated by later studies [2,55].
IS200 is also found on plasmids. For instance, certain lineages of S. abortusovis carry an IS200 element on the virulence plasmid [56]. Hops of IS200 to the pSLT plasmid of S. enterica serovar Typhimurium have been also recovered from stab cultures, confirming that the element can transpose from one replicon to another [3]. Hopping to plasmids might facilitate horizontal transfer of IS200, but evidence for such an event has not been found. IS200 elements are present in strains of E. coli [8] and Shigella [23], and have been also described in Yersinia [31,59]. However, cladograms constructed with IS200 sequences from Salmonella, E. coli, Shigella, and Yersinia match the phylogenetic tree of the Enterobacteriaceae [3,7]. This observation suggests that IS200 elements have not spread among enteric bacteria by horizontal transfer; thus, they must be ancestral components of the enterobacterial genomes [7]. The abundance of IS200 copies in Salmonella contrasts with their erratic occurrence in closely related genera, indicating that the same element can adapt to different lifestyles in different hosts. In addition, the observation that enterics other than Salmonella contain relatively low numbers of IS200 copies is compatible with the hypothesis that stochastic loss of IS200 may have occurred in some lineages.
IS200-like elements have been also found in genera unrelated to enteric bacteria, including Vibrio [6], Enterococcus [18], Clostridium [9], Helicobacter [33], Actinobacillus [29], and even in archaea [57]. Although horizontal transfer cannot be ruled out in all cases, the nucleotide composition of these IS200-like elements is not very different from that of the host genome. Hence, IS200-related transposons may have already existed in remote stages of bacterial evolution, and their endurance can be viewed as a paradox. As parasites, transposons can be expected to be counterselected by their hosts; thus they should be short-lived [53]. One can escape the paradox by admitting that transposons, like all parasites, can increase their evolutionary endurance by attenuating their virulence [21,42]. Natural selection favors parasites with a high reproductive rate, which is the product of two factors: the rate at which new hosts are colonized, and the survival time of the host following infection [42].
Because transposable elements are clonally inherited, host fitness can be considered the main factor in the reproductive rate of the parasite. In natural populations, any strategy that minimizes the risk of causing mutations detrimental to the host will favor maintenance of the element [21]. If this view is correct, transposons with intrinsically low activity will have the highest chances of evolutionary endurance, provided that their copy numbers reach a threshold that prevents stochastic loss.
Practical uses of IS200
If a search for articles on IS200 is carried out in literature databases, more than 100 papers are listed. A few are studies on IS200 organization, function and host range, or descriptions of IS200-related elements in bacterial genera other than Salmonella or Escherichia. The remaining papers, which constitute by far the largest class, deal with the use of IS200 as a molecular marker in medical or veterinary microbiology, especially for epidemiological studies.
The interest in IS200 as a molecular marker for the genus Salmonella relies on two features: its low transposition rate and its widespread distribution. The stability of IS200 copy umber and location allows strain typing, which can be achieved by Southern hybridization or PCR-based methods [16,54]. This allows, for instance, to determine whether two epidemic outbreaks have been caused by the same strain. A further rationale is that IS200 fingerprints can pinpoint evolutionary relatedness among Salmonellae. If two isolates with the same IS200 fingerprint can be viewed as the same strain or "clonal line", divergence in the copy number and distribution of IS200 elements should reflect evolutionary distance. In practice, this approach is often simplistic, especially for Salmonella serovars in which the number of IS200 copies is low. In such cases, IS200 fingerprints may underestimate the polymorphism of natural isolates. Thus, IS200 fingerprinting is often combined with other methods to discriminate among Salmonella serotypes or strains [48,49,50].
Another use of IS200 is the identification of Salmonella in food or in clinical samples of human or veterinary origin. The presence of IS200 in Shigella, a pathogen that causes diseases sharing certain traits with Salmonella infections, ruled out the possibility of using IS200 as a diagnostic probe for the genus Salmonella [23]. However, IS200-based methods have been described for the identification of certain Salmonella serovars. For instance, S. abortusovis carries an IS200 element in a distinct chromosome location. PCR amplification of this serovar-specific IS200 element yields a distinct amplification fragment that is not found with DNAs from other Salmonellae [5]. Serovar-specific IS200 copies have also been described in S. typhi [11] and S. typhimurium [10] and are potentially useful for PCR amplification in diagnostic studies.
Future prospects
The impressive knowledge of transposon biology that has accumulated since the 1970s has been based on the study of elements that are very active and relatively independent from host functions. High activity and (relative) host independence have been key features to permit biochemical and genetic analyses, but may have biased our view of the transposon world. "Desperados" like phage Mu are rare: only a phage can undergo reckless transposition without endangering its chances of survival. A more common type of transposon idiosyncrasy is exemplified by bacterial elements IS50 or IS10, which restrain their own transposition and their copy number per cell but transpose readily under favorable circumstances [15,22,39]. However, less active elements, e.g., IS200 and many eukaryotic transposons, are also common, and their study may uncover novel reactions, rules, and strategies. Of course, working on transposons that rarely transpose will always be difficult, and classical genetic analysis may be severely limited, if not useless. In contrast, the biology of restrained transposons may greatly benefit from fields other than genetics. For instance, genome sequencing programs can identify putative transposons and detect their variants. Serial analysis of gene expression may also identify active copies and conditions that enhance their expression. Of course, function identification and characterization will ultimately rely on biochemistry, and the analysis of multi-component, slow-acting or sloppy transposases may represent a challenge. However, in the last decade, sophisticated biochemical approaches have proven useful to address difficult or elusive aspects of transposase-catalyzed reactions [30,32,37,40,61,62], suggesting that many aspects of transposon complexity are amenable to biochemical dissection.
Acknowledgements. A stay of D.C. at the University of Seville was supported by a Spanish-Italian Collaborative Grant (Acción Integrada HI-2001-0052). A copy of Ken Haack's Ph.D. Thesis was kindly provided by John Roth. We also thank John Roth and Ken Haack for discussions, and for communication of unpublished data.
References
1. Barnes WM (1979) Construction of an M13 histidine-transducing phage: a single-stranded cloning vehicle with one EcoRI site. Gene 5:127-139 [ Links ]
2. Beuzón CR, Casadesús J (1997) Conserved structure of IS200 elements in Salmonella. Nucleic Acids Res 25:1355-1361 [ Links ]
3. Beuzón CR, Casadesús J (1997) Cloning with Mud-P22 hybrid prophages: physical mapping of IS200 elements on the chromosome of Salmonella typhimurium LT2. Mol Gen Genet 256:586-588 [ Links ]
4. Beuzón CR, Marqués S, Casadesús J (1999) Repression of IS200 transposase synthesis by RNA secondary structures. Nucleic Acids Res 27:3690-3695 [ Links ]
5. Beuzón CR, Schiaffino A, Leori G, Cappucinelli P, Rubino S, Casadesús J (1997) Identification of Salmonella abortusovis by PCR amplification of a serovar-specific IS200 element. Appl Environ Microbiol 63:2082-2085 [ Links ]
6. Bik EM, Gouw RD, Mooi FR (1996) DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol 34:1453-1461 [ Links ]
7. Bisercic M, Ochman H (1993) The ancestry of insertion sequences common to Escherichia coli and Salmonella typhimurium. J Bacteriol 175:7863-7868 [ Links ]
8. Bisercic M, Ochman H (1993) Natural populations of Escherichia coli and Salmonella typhimurium harbor the same classes of insertion sequences. Genetics 133:449-454 [ Links ]
9. Brynestad S, Synstad B, Granum PE (1997) The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning isolates. Microbiology 143:2109-2115 [ Links ]
10. Burnens AP, Stanley J, Sack R, Hunziker P, Brodard I, Nicolet J (1997) The flagellin N-methylase gene fliB and a serovar-specific IS200 element in Salmonella typhimurium. Microbiology 143:1539-1547 [ Links ]
11. Calva E, Ordoñez LG, Fernandez-Mora M, Santana FJ, Bobadilla M, Puente JL (1997) Distinctive IS200 insertion between gyrA and rcsC genes in Salmonella typhi. J Clin Microbiol 35:3048-3053 [ Links ]
12. Casadesús J, Naas T, Garzón A, Arini A, Torreblanca J, Arber W (1999) Hotspot absence: a constraint for IS30 transposition in Salmonella typhimurium. Gene 238:231-239 [ Links ]
13. Casadesús J, Roth J R (1989) Transcriptional occlusion of transposon targets. Mol Gen Genet 216:204-209 [ Links ]
14. Casadesús J, Roth J R (1989) Absence of insertions among spontaneous mutants of Salmonella typhimurium. Mol Gen Genet 216:210-216 [ Links ]
15. Chandler M, Mahillon J (2002) Insertion sequences revisited. In: Berg DE, Howe MM (eds) Mobile DNA II. American Society for Microbiology, Washington DC, pp. 305-366 [ Links ]
16. Crichton PB, Old DC, Murphy HF (1998) Distribution of insertion sequence IS200 among different clonal lines of the related Salmonella serotypes Livingstone and Eimsbuettel. J Med Microbiol 47:791-797 [ Links ]
17. Cullum J (1985) Insertion sequences. In: Scaife J, Galizzi A (eds) Genetics of bacteria. Academic, New York, pp 85-101 [ Links ]
18. Dahl KH, Lundblad EW, Rokenes TP, Olsvik O, Sundsfjord A (2000) Genetic linkage of the van2b gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiology 146:1469-1479 [ Links ]
19. Daniell E, Roberts R, Abelson J (1972) Mutations in the lactose operon caused by bacteriophage Mu. J Mol Biol 69:1-8 [ Links ]
20. DeBoy RT, Craig NL (2000) Target site selection by Tn7: att Tn7 transcription and target activity. J Bacteriol 182:3310-3313 [ Links ]
21. Doolittle WF, Kirkwood TBL, Dempster MAH (1984) Selfish DNAs with self-restraint. Nature 307:501-502 [ Links ]
22. Galas DJ, Chandler MJ (1989) Bacterial insertion sequences. In: Berg DE, Howe MM (eds) Mobile DNA. American Society for Microbiology, Washington DC, pp. 109-162 [ Links ]
23. Gibert I, Barbé J, Casadesús J (1990) Distribution of insertion sequence IS200 in Salmonella and Shigella. J Gen Microbiol 136:2555-2560 [ Links ]
24. Gibert I, Carroll K, Hillyard DR, Barbé J, Casadesús J (1991) IS200 is not a member of the IS600 family of insertion sequences. Nucleic Acids Res 19:1343 [ Links ]
25. Gottesman MM, Rosner JL (1995) Acquisition of a determinant for chloramphenicol resistance by bacteriophage lambda. Proc Natl Acad Sci USA 72:5014-5018 [ Links ]
26. Greeb J, Atkins J, Loper JC (1971) Histidinol dehydrogenase (hisD) mutants of Salmonella typhimurium. J Bacteriol 106:421-431 [ Links ]
27. Haack KR, Roth JR (1995) Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics 141:1245-1252 [ Links ]
28. Hartman PE, Hartman Z, Stahl RC, Ames BN (1971) Classification and mapping of spontaneous and induced mutations in the histidine operon of Salmonella. Adv Genet 16:1-34 [ Links ]
29. Hayashida H, Hotokezaka H, Ohara N, Kimura M, Takagi O, Yamada T (1996) Molecular analysis of a new insertion sequence from Actinobacillus (Haemophilus) actinomycetemcomitans FDC Y4. Microbiology 42:2449-2452 [ Links ]
30. Hickman AB, Li Y, Matthew SV, May EW, Craig NL, Dyda F (2000) Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol Cell 5:1025-1034 [ Links ]
31. Hu P, Elliott J, McCready P, Skowronski E, Garnes J, Kobayashi A, Brubaker RR, Garcia E (1998) Structural organization of virulence-associated plasmids in Yersinia pestis. J Bacteriol 180:5192-5202 [ Links ]
32. Kennedy AK, Haniford DB, Mizuuchi K (2000) Single active catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothiotate stereoselectivity. Cell 101:295-305 [ Links ]
33. Kersulyte D, N. S. Akopyants NS, Clifton SW, Roe BA, Berg DE (1998) Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223:175-186 [ Links ]
34. Lam S, Roth JR (1983) IS200: a Salmonella-specific insertion sequence. Cell 34:951-960 [ Links ]
35. Lam S, Roth JR (1983) Genetic mapping of IS200 copies in Salmonella typhimurium strain LT2. Genetics 105:801-811 [ Links ]
36. Lam S, Roth JR (1986) Structural and functional studies of insertion element IS200. J Mol Biol 187:157-167 [ Links ]
37. Lewis RA, Grindley ND (1997) Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol Microbiol 25:517-529 [ Links ]
38. Loper JC, Grabnar M, Stahl RC, Hartman Z, Hartman PE (1964) Genes and proteins involved in histidine biosynthesis in Salmonella. Brookhaven Symp Biol 17:15-50 [ Links ]
39. Mahillon J, Chandler M (1998) Insertion sequences. Microbiol Mol Biol Rev 62:725-774 [ Links ]
40. Manna D, Higgins NP (1999) Phage Mu transposon immunity reflects supercoil domain structure of the chromosome. Mol Microbiol 32:595-606 [ Links ]
41. Matsutani S , Othsubo H, Maeda Y, Othsubo E (1987) Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol 196:445-455 [ Links ]
42. Maynard-Smith J (1989) Evolutionary Genetics. Oxford, England: Oxford University Press [ Links ]
43. McCarthy JEG, Brimacombe R (1994) Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet 10:402-407 [ Links ]
44. Meyer J, Iida S, Arber W (1980) Does the insertion element IS1 transpose into preferentially into A+T-rich DNA segments? Mol Gen Genet 178:471-473 [ Links ]
45. Miko A, Guerra B, Schroeter A, Dorn C, Helmuth R (2002) Molecular characterization of multiresistant d-tartrate-positive Salmonella enterica serovar paratyphi isolates. J Clin Microbiol 40:3184-3191 [ Links ]
46. Nevers P, Saedler H (1977) Transposable genetic elements as agents of gene instability and chromosome rearrangements. Nature 268:109-115 [ Links ]
47. O'Reilly C, Black GW, Laffey R, McConnell DJ (1990) Molecular analysis of an IS200 insertion in the gpt gene of Salmonella typhimurium. J Bacteriol 172:6599-6601 [ Links ]
48. Old DC, Crichton PB, Taylor A, Mather H (2001) An attempt to identify the evolutionary origin of a novel serotype of Salmonella enterica isolated from harbour porpoises. J Med Microbiol 50:415-420 [ Links ]
49. Olsen JE, Skov MN, Threlfall EJ, Brown DJ (1994) Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field electrophoresis and RFLP typing. J Med Microbiol 40:15-22 [ Links ]
50. Olsen JE, Skov MN, Angen O, Threlfall EJ, Bisgaard M (1997) Genomic relationships between selected phage types of Salmonella enterica subsp. enterica serotype Typhimurium defined by ribotyping, IS200 typing, and PFGE. Microbiology 143:1471-1479 [ Links ]
51. Raabe T, Jenny E, Meyer J (1988) A selection cartridge for rapid detection and analysis of spontaneous mutations including insertions of transposable elements in Enterobacteriaceae. Mol Gen Genet 215:176-180 [ Links ]
52. Roth JR, Antón DN, Hartman P (1966) Histidine regulatory mutants of Salmonella typhimurium. I. Isolation and general properties. J Mol Biol 22:305-323 [ Links ]
53. Roth JR, Schmid MB (1981) Arrangement and rearrangement of the bacterial chromosome. Stadler Genet Symp 13:53-70 [ Links ]
54. Rubino S, Muresu E, Solinas M, Santona M, Paglietti B, Azara A, Schiaffino A, Santona A, Maida A, Cappuccinelli P (1998) IS200 fingerprint of Salmonella enterica serotype Typhimurium human strains isolated in Sardinia. Epidemiol Infect 120:215-222 [ Links ]
55. Sanderson KE, Sciore P, Liu SL, Hessel A (1993) Location of IS200 on the genomic cleavage map of Salmonella typhimurium LT2. J Bacteriol 175:7624-7628 [ Links ]
56. Schiaffino A, Beuzón CR, Uzzau S, Leori G, Cappuccinelli P, Casadesús J, Rubino S (1996) Strain typing with IS200 fingerprints in Salmonella abortusovis. Appl Environ Microbiol 62:2375-2380 [ Links ]
57. Schnabel H, Palm P, Dick K, Grampp B (1984) Sequence analysis of the insertion element ISH1-8 and associated structural changes in the genome of phage PH of the archaebacterium Halobacterium halobium. EMBO J 3:1717-1722 [ Links ]
58. Simon R, Hötte B, Klauke B, Kosier B (1991) Isolation and characterization of insertion sequence elements from Gram-negative bacteria using new broad-host-range, positive selection vectors. J Bacteriol 173:1502-1508 [ Links ]
59. Simonet M, Riot B, Fortineau N, Berche P (1996) Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect Immun 64:375-379 [ Links ]
60. Stalder R, Arber W (1989) Characterization of in vitro constructed IS30-flanked transposons. Gene 76:187-193 [ Links ]
61. Turlan C, Ton-Hoang B, Chandler M (2000) The role of tandem IS dimers in IS911 transposition. Mol Microbiol 35:1312-1325 [ Links ]
62. Williams TL, Jackson EL, Carritte A, Baker TA (1999) Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev 13:2725-2737 [ Links ]
63. Zerbib D, Gamas P, Chandler M, Prentki P, Bass S, Galas D (1985) Specificity of insertion of IS1. J Mol Biol 185:517-524 [ Links ]