Meu SciELO
Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
International Microbiology
versão impressa ISSN 1139-6709
INT. MICROBIOL. vol.8 no.3 Set. 2005
| RESEARCH ARTICLE | |||
|
| |||
| Biofilm formation in spent nuclear fuel pools and bioremediation of radioactive water
Summary. Microbiological studies of spent nuclear fuel pools at the Cofrentes Nuclear Power Plant (Valencia, Spain) were initiated to determine the microbial populations in the pools' water. Biofilm formation at the nuclear power plant facilities and the potential use of those microbial populations in the bioremediation of radioactive water were also studied. Biofilm formation was analyzed by immersing different austenitic stainless steel coupons (UNS S30400, UNS S30466, UNS S31600), as well as balls of stainless steel (UNS S44200) and titanium (99.9%) in a spent nuclear fuel pool (under static and dynamic conditions) for 34 months. Epifluorescence microscopy and scanning electron microscopy revealed that biofilm formed on the samples, in spite of the radioactive and oligotrophic conditions of the water. Based on standard culture methods and sequencing of 16S rDNA fragments, 57 bacteria belonging to α-, β-, and γ-Proteobacteria, Firmicutes and Actinobacteridae were identified in the biofilms. The radioactivity of the biofilm was measured using γ-ray spectrometry, which revealed that biofilms were able to retain radionuclides, especially 60Co. Using metallic materials to decontaminate radioactive water could become a new approach for bioremediation. [Int Microbiol 2005 8(3):223-230] Key words: nuclear power plant · spent nuclear fuel pool · radioactive water · biofilms · bioremediation | ||
| |||
Formación de biopelículas en piscinas de combustible nuclear gastado y biorremediación de aguas radiactivas Resumen. Los estudios microbiológicos de las piscinas de combustible nuclear gastado en la Central Nuclear de Cofrentes (Valencia, España) se iniciaron para determinar las poblaciones microbianas del agua de dichas las piscinas. También se estudió la formación de biopelículas en las instalaciones de la central nuclear y del posible uso de esas poblaciones microbianas para la biorremediación de aguas radiactivas. Para analizar la formación de las biopelículas en una piscina de combustible nuclear gastado, se sumergieron durante 34 meses (en condiciones estáticas y dinámicas) cupones de acero inoxidable austenítico (UNS S30400, UNS S30466, UNS S31600), así como ovillos de acero inoxidable (UNS S44200) y titanio (99.9%). La observación al microscopio de epifluorescencia y al microscopio electrónico de barrido reveló la formación de biopelículas sobre las muestras a pesar del carácter radioactivo y oligotrófico del agua. De acuerdo con los métodos estándar de cultivo y secuenciación de los fragmentos del 16S rDNA, en las biopelículas se identificaron 57 bacterias diferentes pertenecientes a los grupos α-, β-, y γ-Proteobacteria, Firmicutes y Actinobacteridae. La radiactividad de las biopelículas se midió por espectrometría gamma y esta técnica demostró que las biopelículas son capaces de retener radionucleidos, especialmente 60Co. El empleo de materiales metálicos para descontaminar aguas radiactivas podría ser un nuevo método de biorremediación. [Int Microbiol 2005 8(3):223-230] Palabras clave: central nuclear · piscina de combustible nuclear gastado · agua radiactiva · biopelículas · biorremediación | Formação de biofilmes em piscinas de combustível nuclear usado e bioremediação de águas radioativas Resumo. Os estudos microbiológicos das piscinas de combustível nuclear gasto na Central Nuclear de Cofrentes (Valência, Espanha) se iniciaram para determinar as povoações microbianas da água dessas piscinas. Também se estudou a formação de biofilmes nas instalações da central nuclear e o possível uso dessas povoações microbianas para a bioremediação de águas radiativas. Para analisar a formação dos biofilmes em uma piscina de combustível nuclear usado, durante 34 meses (em condições estáticas e dinâmicas) se mantiveram ali submergidos cupons de aço inoxidável austenítico (UNS S30400, UNS S30466, UNS S31600), assim como novelos de aço inoxidável (UNS S44200) e titânio (99.9%). A observação ao microscópio de epifluorescéncia e ao microscópio eletrônico de varredura revelou a formação de biofilmes sobre as amostras a pesar do caráter radioativo e oligotrófico da água. De acordo com os métodos standard de cultivo e secuenciación dos fragmentos do 16S rDNA, nas biopelículas se identificaram 57 bactérias diferentes pertencentes aos grupos α-, β-, e γ-Proteobacteria, Firmicutes e Actinobacteridae. A radiatividade das biopelículas se mediu por espectrometria gamma, que demonstrou que os biofimes podiam reter radionucleidos, especialmente 60Co. O emprego de materiais metálicos para descontaminar águas radiativas poderia ser um novo método de bioremediação. [Int Microbiol 2005 8(3):223-230] Palavras chave: central nuclear · piscina de combustível nuclear usado · água radiativa · biofilmes · bioremediação |
Introduction
In order to generate electrical energy in light-water nuclear power plants (NPPs), enriched uranium is used as fuel. Uranium oxide pellets are placed inside hollow bars of zircaloy-2 (a zirconium alloy), forming fuel elements that are positioned in the core so that nuclear fission can be carried out. However, after a certain time, the fuel elements can no longer effectively produce energy. These "spent" elements need to be replaced in a process known as refueling. Since the spent nuclear fuel (SNF) continues to be thermally and radiologically active, it needs to be stored in pools. Thus, it must be moved from the reactor's pools (located in the reactor building) to the SNF pools (located in the fuel building) by means of a fuel transference pipe. The water from the pools has two functions: it protects against radiation, and contributes to cooling the fuel. The quality of the water must comply with strict requirements of purity and clarity so that maintenance personnel can correctly place the spent fuel elements on submerged racks.
Nuclear pools were initially designed to store elements generated over 20 years, a time period considered reasonable before deciding how the spent fuel would finally be treated. However, at many NPPs, including the one located in Cofrentes, Valencia, Spain, the need to increase the capacity of the pools and to store the spent fuel for the lifetime of the power plant, which is approximately 40 years, is now seen as more convenient. To do this, it is necessary to double the initial capacity of the pools, which has been made possible by implementing racks that have a greater capacity [Rebollo C, Arana S, Cerezo I (1995) Integrated project for increasing the capacity of spent fuel pools at Cofrentes NPP. In: Sociedad Nuclear Española (ed) XXI Reunión Anual, Madrid, Spain, pp 239-257]. This reracking means that the quality of water must meet strict physico-chemical conditions.
Given the oligotrophic and radiological character of the water, and from a strictly microbiological point of view, the active microbial activity found in the pools is very unusual. Bacteria belonging to very diverse phylogenetic groups have been identified, not only in the water itself, but also in the austenitic stainless steel walls cladding the pools. In early assays, in which stainless steels were submerged in the pools, bacteria rapidly developed biofilms on the submerged material [27]. This led us to focus our research on the possibility of using these biofilms for the bioremediation of nuclear water.
Materials and methods
Spent nuclear fuel pool and water analyses. Studies were carried out in the west SNF pool of the Cofrentes NPP (Valencia, Spain). The nuclear pools have thick concrete walls clad in austenitic stainless steel and are permanently filled with water. The water in the SNF pools is cooled and purified in a closed-loop system that includes filtration and demineralization processes that ensure the high-purity of the water. Physical-chemical and radiochemical analyses of the water were carried out once a week in the SNF pool, as described previously [6].
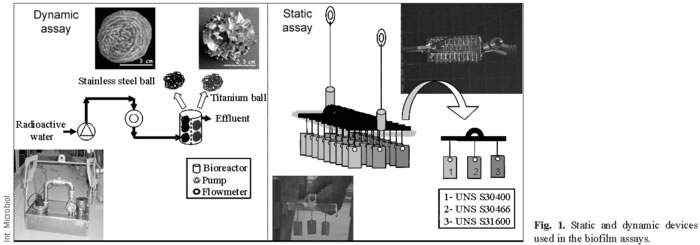
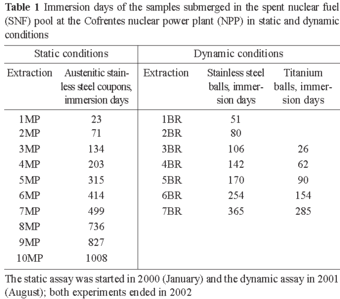
Metal coupons and biofilm tests. Two different assays using metallic materials were developed for use in the SNF pool, under static and dynamic conditions. In the first, static assay, coupons of three different types of austenitic stainless steel in the as-received state, with surfaces measuring 14 (UNS S30400), 13.5 (UNS S31600), and 15.5 cm2 (UNS S30466-borated), were used. The coupons were separately installed on the racks in groups of three; each group consisting of one coupon made of each material (Fig. 1). The composition of theses austenitic stainless steel coupons was similar or identical to the cladding surfaces of different facilities of the SNF pools. In the second assay, carried out under dynamic conditions (capacity of the pump: 2.3 m3·h-1), a stainless steel bioreactor was used that contained balls of two different materials: stainless steel (≈UNS S44200), weighing 20 g each, and titanium (99.9%), weighing 5 g each (Fig. 1). In both assays, the materials were submerged at a depth of 3 m in the west SNF pool at the Cofrentes NPP. The materials selected were non-corrosive and non-degradable in this environment. They were degreased using acetone in an ultrasound bath and sterilised before being submerged. After different immersion times (Table 1) (1008 days under static conditions, and 365 days under dynamic ones), the coupons and balls were carefully removed in a sterile environment and transported to the laboratory in test tubes filled with sterile water. In all cases, the samples were extracted from the NPP only when the radioactivity levels permitted it.
Epifluorescence microscopy. The microbiological quality of the water was analyzed as described for membrane filters [6]. The biofilms formed under static and dynamic conditions were visualized by epifluorescence microscopy (Axioskop 2, Zeiss, Germany) using the LIVE/DEAD BacLight Viability Kit (L-7012, Molecular Probes Inc.) as previously described [27]. Microphotographs were obtained using an Olympus Digital Camera C-3030 Zoom. Biofilm formation on the metal coupons (UNS S30400) under static conditions was evaluated until the 134th day of immersion. After that, the high levels of radioactivity no longer allowed the coupons to be extracted from the NPP. This problem did not arise under dynamic conditions. In order to transport the metallic materials to the biodeterioration laboratory, small fragments of the metallic balls (measuring less than a 1 cm2) were cut and maintained in Milli-Q sterile water.
Scanning electron microscopy. Scanning electron microscopy (SEM) analyses were carried out on UNS S31600 coupons (13.5 cm2) for the static experiment, and on balls (measuring approximately 1 cm2), as described above, for the dynamic experiment. The samples were washed in sodium cacodylate 0.01 M buffer and then fixed with 2.5% glutaraldehyde in sodium cacodylate 0.01 M at 4ºC for 2-3 h. After that, the samples were dehydrated using a series of acetone-water washes (20%, 40%, 60%, 80%) and submerged in these solutions for 30 min at 4ºC. Finally, the samples were maintained in a 100% acetone solution at 4ºC. The specimens were further processed using the critical point procedure (CPD 020, Balzers Union), sputter-coated with gold (SCD 004, Balzers Union), and observed under a scanning electron microscope (DSM 960, Zeiss) operated at 15 kV accelerating voltage.
Bacterial isolation and DNA extraction. Viable and culturable microorganisms formed sessile population on samples under static and dynamic conditions were isolated by one of three different methods. Method 1 was used only for metal coupons of UNS S30466 until day 203. The coupons were then removed from the pool and incubated in nutrient broth (Nutrient Broth no. 2, Oxoid) at 30ºC for approximately 72 h. Afterwards, to isolate the large number of species, 100 µl of the bacterial suspension was plated on three different solid media, as described previously [27]. Cultures were incubated at 30ºC until colonies appeared. Colonies growing on the plates were picked and purified several times by restreaking. Method 2 was used for coupons of the static assay (UNS S30400, UNS S31600, and UNS S30466) after 203 immersion days, at which point the high levels of radiation no longer permitted removal of the coupons from the NPP. The coupons were washed with sterile water, and the biofilm was removed and transferred to 100 ml of sterile water before being submitted to sonication for 15 min. A 25-ml portion of this half-diluted water was filtered through 0.45-µm filters (HAWP04700, MF membrane filters, Millipore). Filters were incubated inside the NPP in different solid media: TSB (Tryptone Soya Broth, Oxoid; supplemented with agar (UPS), purissimum, Panreac), STC and R2 as in [27]. Cultures were incubated at 30ºC until colonies appeared. Colonies growing on the plates were picked and purified several times by restreaking. In method 3, biofilms formed under dynamic conditions were removed from the samples using an ultrasound bath for 15 min in 250 ml of deionized sterile water. After sonication, 50 ml of a 1:10 dilution was filtered through 0.45-µm filters (MF™Membrane filters, Millipore) as above, and the filters were plated onto different solid media (NA, BHI, TSB, STC, R2) as described previously [27]. Due to the highs levels of radiation in all cases (up to 2030 Bq·cm-2), the filters were incubated at 30ºC until colonies appeared in the NPP. Colonies growing on the plates were picked inside the NPP and purified by restreaking several times on the same culture media. Genomic DNAs of the isolated microorganisms were then extracted using Prep Man Ultra (PE Applied Biosystems), according to the protocol given by the manufacturer, or freeze-thawed (-20ºC, +60ºC). Colonies of different morphologies isolated by the above-described methods were picked from each plate, purified, and maintained at -20ºC using the kit (KPX007, Microkit Iberica).
Polymerase chain reaction and denaturing gradient gel electrophoresis analyses. 16S rDNA fragments were amplified by "touchdown" PCR using the universal primers of Escherichia coli 5F (containing a 40-base GC-clamp at its 5´ end,) and 531R. The annealing temperature was lowered from 50ºC to 40ºC over 20 cycles. The following 20 cycles were set at 43ºC. Each cycle consisted of an initial denaturing step lasting 45 s at 94ºC, and a final elongation step lasting 45 s at 72ºC. In all these reactions, a PCR-master kit (Roche) was used. PCRs were carried out in a GeneAmp PCR System 2400 (Perkin Elmer, Norwalk, Conn.). Amplification products were analyzed by electrophoresis as described earlier [27]. Ten µl of the amplified fragments were analyzed by denaturing gradient gel electrophoresis (DGGE), with a 30-60% denaturant gradient (100% denaturant contains 7 M urea and 40% [v/v] formamide). DGGE and preincubation of the gel were carried out as previously reported [27].
Sequencing and phylogenetic identification. Bacteria were identified by sequencing 500 bp using MicroSeq 500 16S rDNA or the BigDye Terminator v1.1 Cycle Sequencing kit (L-7012, PE Applied Biosystems). Sequences were resolved in an ABI PRISM 310 Genetic Analyzer following the manufacturer's instructions. The sequences obtained were compared directly to all the known sequences deposited in the NCBI databases using the basic local alignment search tool Blastn. Sequences were aligned using CLUSTAL_X software, version 1.81 [30]. Phylogenetic and molecular evolutionary analyses were conducted using MEGA (Molecular Evolutionary Genetics Analysis) version 2.1 [19]. Phylogenetic trees were constructed using the neighbor-joining method with the Jukes-Cantor model [17] and maximum parsimony methods. A total of 1000 bootstrapped replicate resampling data sets were generated.
Nucleotide sequence accession numbers. The sequences obtained from microorganisms forming biofilms in the SNF pools were deposited in GenBank under the accession numbers AY599885 to AY599892, AY894720 to AY894731, and AY791999 to AY792035. Additionally, the sequences of microorganisms isolated from the walls of the reactor's pools were assigned the accession numbers AF397059 to AF397063, AY479983, and AY479984, and those from the SNF pool water; AY509953 to AY509973.
Accumulation of radionuclides in the biofilms. The biofilm formed on the coupons under static conditions were removed with 100 ml deionized sterile water solution in an ultrasonic cleaner in order to analyze the water's radioactivity, which is an indirect measurement of the radioactivity in the biofilm. In the dynamic assay, the metallic balls were directly introduced into the detector after being washed in sterilized water. In case the detector was saturated, the metallic balls were cut in two or more pieces. One of them was weighed before being introduced in the detector in order to obtain a measure of residual radioactivity using the same protocol.
Manipulation of samples and waste management. All samples obtained in this study were collected and manipulated following the recommendations of the Nuclear Security Council to prevent radiological contamination [8]. The materials used in this study and the waste products generated were treated as radioactive waste.
Results
Water quality. The concentration of nitrates, sulfates, and chlorides in water treated in the closed-loop system was typically within the 0.45-0.59 µg/l range, the water conductivity was 0.8-1.5 µScm-1, pH values ranged from 5.0 to 6.5, the temperature was between 25 and 30ºC, and the TOC values ranged from 0.01 to 0.61 ppm. These data are in accordance with the recommended values given for the NPP, and indicate the oligotrophic character of the water. The radiochemical analysis of the water by γ-spectrometry is shown in Fig. 2. The high peak matches with the refueling period. The most common radionuclides found in the water were 60Co, 137Cs, 134Cs, 54Mn, and 65Zn.
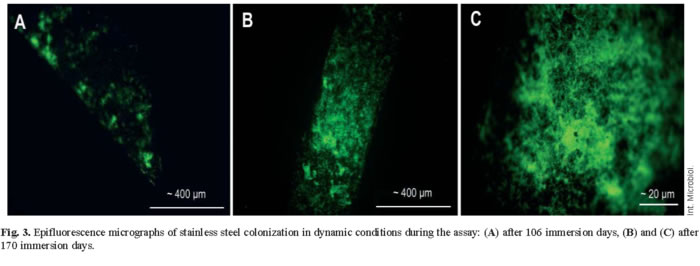
Microscopy analysis of biofilms. Epifluorescence microscopy studies revealed that both in static and dynamic conditions, the metallic materials were progressively colonised with time (Fig. 3). In all materials the colonization mainly consisted in filamentous and long rod-shaped bacteria. Even though a high number of dead cells were found in the oldest biofilms, the living and metabolically active bacteria still outnumbered the dead ones in spite of the high levels of radiation.
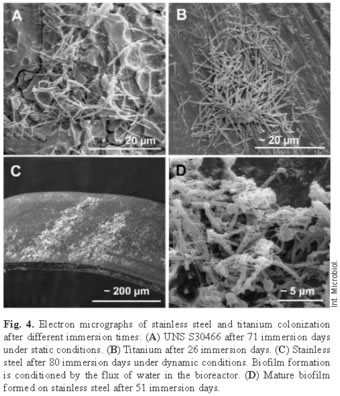
Scanning electron microscopy confirmed the epifluorescence results showing that there was effectively bacterial colonization and that the microorganisms in the biofilms were diverse. In static conditions, complex biofilm was formed on stainless steel after 71 days in this radioactive and oligotrophic environment (Fig. 4). In dynamic conditions, the biofilm was observed after 51 immersion days, as well as the flux influence in the biofilm structure. On the titanium, the colonization was apparent after 26 immersion days. It was at this point that the different types of microorganisms appeared, as on the stainless steel. Differences in the colonization could be seen on the surface morphology of the titanium material. On the rough or obverse side, the colonization was more intense; it even covered completely the surface of the material. This side had a few colonies that were very large; while the smooth or reverse side had more colonies that were smaller.
Bacterial isolation and identification. From the filters cultured on different solid media, 283 colonies were isolated: 94 from static assays, and 146 from dynamic ones. The PCR products of the total number of microorganisms were analyzed using DGGE, thus allowing us to differentiate the migration patterns, and making it possible to use it as pre-screening test. From the total of microorganisms, 20 different bacteria were identified in the static conditions, and 37 in the dynamic ones. These bacteria belonged to α-, β-, and γ-Proteobacteria, Firmicutes and Actinobacteridae. On austenitic stainless steels in static conditions, 12 different genera were identified; while on UNS S44200 in dynamic conditions, they were 8 genera and on titanium 7 genera. On UNS S30400 and UNS S31600 stainless steels in static conditions, the bacterial diversity in the biofilm increased with time and reached the highest diversity at 827 immersion days (4 genera), while on UNS S30466, it was at 499 immersion days (3 genera). On UNS S44200 in dynamic conditions, the highest diversity was found after 170 immersion days (6 genera). However, the diversity in the biofilm on titanium was similar regardless of the immersion time, with 4 of 5 genera per analyzed sample. Colonization was different on different materials. In static conditions, Staphylococcus was the only genus present in the biofilm of all types of austenitic stainless steel, regardless the immersion time. In dynamic conditions, the most represented genera in the biofilm developing on the stainless steel balls were Bacillus and Stenotrophomonas, whereas in titanium biofilm, they were Ralstonia (=Wautersia, =Cupriavidus) and Mycobacterium (Fig. 5).
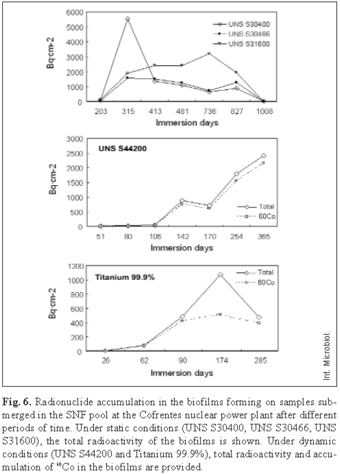
Biofilm radioactivity. The analysis of the biofilms developing on samples submerged in the pools during the studies showed that radionuclides present in water accumulated on the surface of the metallic samples. The accumulation was different depending on the material. The main radionuclides accumulated in the biofilm on stainless steel in both static and dynamic conditions were 60Co, 65Zn and 54Mn, whereas a few radionuclides of other varieties could be detected. The most abundant radionuclides in static conditions on austenitic stainless steel correspond to 60Co, 65Zn, 51Cr, but in dynamic conditions on UNS S44200, 51Cr disappeared, and 54Mn appeared in the biofilms. In titanium biofilms, other radionuclides appeared such as 59Fe, 95Nb, and 65Zn. 60Co was the most common radionuclide in the biofilm and the accumulation curves were defined mainly by the accumulation pattern of this radionuclide (Fig. 6).
In static conditions, the maximum radioactivity rate on the coupons at 315 days was observed at 5521 Bq·cm-2 for UNS S30400, and 1,560 Bq·cm-2 for UNS S30466. In the case of UNS S31600, the maximum was registered after 736 immersion days with a radioactivity rate of 3163 Bq·cm-2. In dynamic conditions on UNS S44200, the radioactivity increased as of 106 immersion days, and at a maximum of 142 days, the radionuclides rose to 895 Bq·cm-2. After this point, the radioactivity decreased slightly, and then increased again at 170 immersion days, reaching its peak at 365 immersion days with values at 2408 Bq·cm-2. On titanium, radioactivity accumulates quicker than on any of the stainless steel types in both static and dynamic conditions. The peak was reached at 174 immersion days with values at 1075 Bq·cm-2.
Discussion
This research shows that microorganisms belonging to different phylogenetic groups present in the water of SNF pools are viable and can grow and form biofilms on stainless steel and titanium, in spite of the oligotrophic and radioactive environment. The average number of total microbial cells counted by epifluorescence microscopy in the nuclear fuel pools was 103 cells·ml-1. When culturable methods were used, the range was between 0.41 and 1.77 CFU·ml-1 [7]. In all cases, culturable microorganisms were about 0.1% of the total microorganisms counted by epifluorescence microscopy. Similar values have also been found by other researchers [26] and seem to be a standard value in SNF storage waters.
Sequencing techniques provided an analysis of the bacterial 16S rDNA gene composition present in the biofilms of stainless steels and titanium samples. The bacteria in the biofilms belonged to five different phylogenetic groups (α-, β-, and γ-Proteobacteria, Actinobacteridae, and Firmicutes) and were found on both materials, but some species could only be isolated from one of them. Among the isolated bacteria, gram-positive and gram-negative were found. The percentages of gram-positive and gram-negative were similar on different materials, but the numbers of gram-negative bacteria in both the biofilms and the water of the SNF pools [6] were higher than those found in other studies in radioactive environments [15]. Such differences might be due either to different culture conditions or to the fact that biofilms would function as a radiological protection for the microorganisms that make up it. Although the radioresistance of bacteria isolated in this research has not been tested, other bacteria isolated in the same SNF pools belonging to similar genera presented D10 values of about 1.4 kGy (data not published). Moreover, microorganisms were growing in the culture media on filters with radioactivity rates of up to 2030 Bq·cm-2.
The γ-spectrometry analyses during the assays showed that the radioactivity due to radionuclide accumulation in the biofilm increased with time. The increase in the radioactivity of the biofilm was mainly due to the accumulation of 60Co, which is the most abundant radionuclide in the water of the SNF pool. In stainless steel biofilms, the main radionuclides were 60Co, 65Zn, 54Mn, and 51Cr; while in titanium biofilms, they were 59Fe and 95Nb. These differences imply that there is a selective 59Fe and 95Nb accumulation in the titanium biofilms, because both radionuclides are less abundant in the water than 65Zn, 54Mn, and 51Cr. On the other hand, they accumulated more quickly in titanium than in stainless steel during the first immersion period. During longer immersion periods, however, radioactivity accumulation was higher in stainless steels biofilms. The differences between 54Mn accumulation and the maximum biofilm radioactivity in stainless steel and titanium might be due to the existence of different microorganisms with Fe-independent or Mn-energy dependent accumulation. The Mn/Fe ratio is higher in biofilms developing on stainless steel than in those developing on titanium. To explain such a difference, it has been suggested that Mn (II) accumulation (with low Fe) might be a widespread strategy for survival [9].
The differences in bacterial composition of biofilms and in the accumulation processes of radionuclides might account for the differences of radioactivity observed between materials. In titanium biofilm, the high diversity of Ralstonia might be due to the rapid increase in radioactivity. This genus, which has been isolated in radioactive and/or oligotrophic environments before forming biofilms [7,18,22,24], is involved in biomineralization and chemisorption processes forming hydroxides, carbonates [21], metallic radionuclides [20], iron oxides and manganese oxides [5,28]. Burkholderia, which was isolated from the water of SNF pools in a previous study [7], is considered to be an ubiquitous genus of soil bacteria related to PAH-degrading [23], TCE-degrading [4], and herbicide-degrading [16] processes. Both Ralstonia and Burkholderia have been classified by the Microbial Genomics Research, DOE (Department of Energy, USA) [http://www.microbialgenome.org/organisms.shtml] as microorganisms potentially useful for bioremediation of several compounds and elements.
At advanced stages of immersion, the most abundant genus found on the materials tested was Bacillus, especially B. cereus. This genus has an impressive physiological diversity, and its spores are highly resistant to unfavorable conditions. In the case of B. cereus, it has been characterized as a radioresistant bacterium with D10 values of 1.0 kGy [12]. The Bacillus genus is resistant to gamma radiation, UV, H2O2, and desiccation [25]. It has also been linked to the selective accumulation of Mn, Zn, Cs, U, and Co, among others [25, 29]. Moreover, some strains, and especially their spores, can bind irreversibly large amounts of metals, and the presence of spores at advanced stages of colonization might account for the increasing radioactivity in the biofilm.
Other genera predominant in the biofilms were Stenotrophomonas and Mycobacterium. While the latter has not been previously documented in radioactive environments, both genera have been related to degrading processes of organic compounds [11] or hydrocarbons [2]. Some isolates belonging to Stenotrophomonas can resist doses of up to 2.5 kGy of γ-radiation [13]. Nocardia and Staphylococcus, also present in the biofilms, were previously isolated in the water of SNF pools [7, 27] and in other radioactive environments [13]. These are microorganisms with D10 values higher than 0.5 kGy [3] and 2.5 kGy [13], respectively.
Studies in laboratories and in natural environments have shown a high affinity of microbial biomass for actinide elements, heavy metals, and radionuclides [14]. The results suggest that biofilm communities in the SNF pools are directly involved in the accumulation of radionuclides from the water. The two main mechanisms used by the microorganisms to remove the radionuclides are biosorption and bioaccumulation. Biosorption is a metabolically independent physical process at the cell surface, while bioaccumulation is an energy-dependent process involving intracellular accumulation. Most of the radionuclides detected in the biofilm could be related to biosorption processes, especially 60Co (one of the most abundant radionuclides in the water of SNF pools in NPPs with BWR).
Due to the difficulties in developing this type of research in an active NPP, this study is an important contribution to understanding the role of microorganisms in oligotrophic and radioactive water. The microorganisms and the metallic supports could find application in in-situ bioremediation processes at NPPs.
Future prospects. Bioremediation of water contaminated with radionuclides depends mainly on the capacity of the microorganisms to survive in a radioactive environment. To understand the mechanisms that enable the removal of radionuclides from water and their concentration in bacterial cells, it is necessary to identify not only the culturable microorganisms present in the radioactive water, but also those that are non-culturable. The latter could play a major role in biofilms developing on metallic materials in contact with radioactive water. For this reason, more in-depth studies are needed, using molecular biological culture-independent techniques to detect and identify non-culturable bacteria. For the characterization of environmental samples, the most common approach is to combine DGGE, which generates characteristic band patterns, with the cloning and sequencing of amplified fragments of 16S rDNA. Another common technique is fluorescent in situ hybridization (FISH), which, in addition to contributing in identifying the microorganisms, allows their quantification and a study of their distribution in the biofilms. Another approach is the use of microarrays to detect 16S rRNA or its genes. Microarrays are based on the interaction of complementary DNA chains. Specific single-stranded DNA fragments (ssDNA probes) are immobilised on glass slides or silicon chips and then put into contact with ssDNA samples previously marked with fluorochromes, allowing hybridized complementary probes to be detected fluorescently. Given that the position and identity of each one of the probes is known, it is possible to determine which ones are present in the sample. Currently, microarrays are being used to characterize microbial populations and to detect microorganisms in complex mixtures, although they have not been applied yet to the study of radioactive waters of nuclear power plants.
Acknowledgements. We are grateful to the Spanish Ministry of Science and Technology (CICYT-FEDER-2FD97-0530-MAT), to Iberdrola Generación, S.A. (6276-99) for their financial support and to the staff of the Cofrentes Nuclear Power Plant for their technical assistance.
References
1. AWWA-APHA-WEF (1998) Standard methods for the examination of water and wastewater, 20th edn. Amer Publ Health Assoc, Washington DC [ Links ]
2. Andreoni V, Cavalca L, Rao MA, Nocerino G, Bernasconi S, Dell'Amico E, Colombo M, Gianfreda L (2004) Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere 57:401-412 [ Links ]
3. Aziz NH, El-Fouly MZ, Abu-Shady MR, Moussa LAA (1997) Effect of gamma radiation on the survival of fungal and actinomycetal florae contaminating medicinal plants. Appl Radiat Isotop 48:71-76 [ Links ]
4. Bourquin AW, Mosteller DC, Olsen RL, Smith MJ, Reardon KF (1997) Aerobic bioremediation of TCE-contaminated groundwater: bioaugmentation with Burkholderia cepacia PR1301. In: In Situ and On-Site Bioremediation Symposium, Vol 4, Battelle Press, Columbus, OH, pp 513-518 [ Links ]
5. Brooks SC, Herman JS (1998) Rate and exent of cobalt sorption to representative aquifer minerals in the presence of a moderately strong organic ligand. Appl Geochem 13:77-88 [ Links ]
6. Chicote E, García AM, Moreno DA, Sarró MI, Lorenzo PI, Montero F (2005) Isolation and identification of bacteria from spent nuclear fuel pools. J Ind Microbiol Biot 32:155-162 [ Links ]
7. Chicote E, Moreno DA, García AM, Sarró MI, Lorenzo PI, Montero F (2004) Biofouling of the walls of a spent nuclear fuel pool with radioactive ultrapure water. Biofouling 20:35-42 [ Links ]
8. Council Directive 96/29/EURATOM of 13 May 1996. Laying down basic safety standards for the protection of the health of workers and the general public against the dangers arising from ionizing radiation. Official Journal L 159, 29/06/1996, pp 0001-0114 [ Links ]
9. Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Venkateswaran A, Hess M, Omelchenko MV, Kostandarithes HM, Makarova KS, Wackett LP, Frederickson JK, Ghosal D (2004) Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma radiation resistance. Science 306:1025-1028 [ Links ]
10. Debertin K, Helmer RG (1988) Gamma-ray and X-ray spectrometry with semiconductor detectors. Elsevier Science BV, Amsterdam, pp. 258-281 [ Links ]
11. Esteve-Núñez A, Caballero A, Ramos JL (2001) Biological degradation of 2,4,6-trinitrotoluene. Microbiol Mol Biol Rev 65:335-352 [ Links ]
12. Farrag HA, El-Bazza, ZEM, El-Fouly MED, El-Tablawy SYM (2000) Effect of gamma radiation on the bacterial flora of Nigella sativa seeds and its oil constituents. Acta Pharm 50:195-207 [ Links ]
13. Fredrickson JK, Zachara JM, Balkwill DL, Kennedy D, Li S-MW, Kostandarithes HM, Daly MJ, Romine MF, Brockman FJ (2004) Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington State. Appl Environ Microbiol 70:4230-4241 [ Links ]
14. Gadd GM (2004) Microbial influence on metal mobility and application for bioremediation. Geoderma 122:109-119 [ Links ]
15. Gazsó LG (1997) Basic radiation microbiology. In Wolfram JH, Rogers RD, Gazsó LG (eds) Microbial degradation processes in radioactive waste repository and in nuclear fuel storage areas. Kluwer Academic Publishers, Dordrecht, NATO ASI Series 1, Disarmament Technologies, vol 11, pp 93-101 [ Links ]
16. Jacobsen CS (1997) Plant protection and rhizosphere colonization of barley by seed inoculated herbicide degrading Burkholderia (Pseudomonas) cepacia DBO1 (pRO101) in 2,4-D contaminated soil. Plant Soil 189:139-144 [ Links ]
17. Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed), Mammalian protein metabolism. Academic Press, New York, pp 21-132 [ Links ]
18. Kulakov LA, McAlister MB, Ogden KL, Larkin MJ, O'Hanlon JF (2002) Analysis of bacteria contaminating ultrapure water in industrial systems. Appl Environ Microbiol 68:1548-1555 [ Links ]
19. Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245 [ Links ]
20. Lloyd JR, Macaskie LE (2000) Bioremediation of radionuclide-containing wastewaters. In: Lovley DR (ed), Environmental microbe-metal interactions. ASM Press, Washington DC, pp 277-327 [ Links ]
21. Lovley DR, Lloyd JR (2000) Microbes with a mettle for bioremediation. Nature Biotechnol 18:600-601 [ Links ]
22. McAlister MB, Kulakov LA, O'Hanlon JF, Larkin MJ, Ogden KL (2002) Survival and nutritional requirements of three bacteria isolated from ultrapure water. J Ind Microbiol Biot 29:75-82 [ Links ]
23. Mueller JG, Devereux R, Santavy DL, Lantz SE, Willis SG, Pritchard PH (1997) Phylogenetic and physiological comparisons of PAH-degrading bacteria from geographically diverse soils. Anton Leeuw 71:329-343 [ Links ]
24. Nikitin DI, Tashtemirova MA, Pitriuk IA, Sorokin VV, Oranskaia MS, Nikitin LE (1993) High resistance of some oligotrophic bacteria to ionizing radiation. Mikrobiologiia 62:1064-1071 (In Russian) [ Links ]
25. Romanovskaia VA, Rokikto PV, Mikheev AN, Gushcha NI, Malashenko IuR, Chernaia NA (2002) The effect of gamma-radiation and desiccation on the viability of the soil bacteria isolated from the alienated zone around the Chernobyl Nuclear Power Plant. Mikrobiologiia 71:705-712 (In Russian) [ Links ]
26. Santo Domingo JW, Berry CJ, Summer M, Fliermans CB (1998) Microbiology of spent nuclear fuel storage basins. Curr Microbiol 37:387-394 [ Links ]
27. Sarró MI, Moreno DA, Chicote E, Lorenzo PI, García AM, Montero F (2003) Biofouling on austenitic stainless steels in nuclear spent fuel pools. Mater Corros 54:535-540 [ Links ]
28. Saunders JA, Pritchett MA, Cook RB (1997) Geochemistry of biogenic pyrite and ferromanganese coatings from a small watershed: a bacterial connection? Geomicrobiol J 14:203-217 [ Links ]
29. Selenska-Pobell S, Panak P, Miteva V, Boudakov I, Bernhard G, Nitsche H (1999) Selective accumulation of heavy metals by three indigenous Bacillus strains, B. cereus, B. megaterium and B. sphaericus from drain waters of a uranium waste pile. FEMS Microbiol Ecol 29:59-67 [ Links ]
30. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876-4882 [ Links ]