Meu SciELO
Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista de Osteoporosis y Metabolismo Mineral
versão On-line ISSN 2173-2345versão impressa ISSN 1889-836X
Rev Osteoporos Metab Miner vol.11 no.4 Madrid Nov./Dez. 2019 Epub 06-Abr-2020
https://dx.doi.org/10.4321/s1889-836x2019000400004
ORIGINALS
Functional impact of sclerostin gene polymorphisms on DNA methylation and gene expression
1Internal Medicine Unit. Marqués de Valdecilla University Hospital. Valdecilla Health Research Institute (IDIVAL). University of Cantabria. Santander (Spain).
2Department of Legal Medicine. School of Medicine. University of Cantabria. Santander (Spain).
3Traumatology Unit. Marqués de Valdecilla University Hospital. Valdecilla Health Research Institute (IDIVAL). University of Cantabria. Santander (Spain).
4University Institute of Oncology of the Principality of Asturias (IUOPA). Institute of Health Research of the Principality of Asturias - Central University Hospital of Asturias (ISPA-HUCA). Foundation for Biosanitary Research of Asturias (FINBA). Asturias (Spain).
5Cancer Epigenetics Laboratory. University Institute of Oncology of the Principality of Asturias (IUOPA). Nanomaterials and Nanotechnology Research Center - Higher Council for Scientific Research (CINN-CSIC). Oviedo University. Asturias (Spain).
Introduction:
Several genome-wide association studies (GWAS) and others which focused on the sclerostin gene (SOST) have found that some SOST polymorphisms are associated with bone mass and risk of fractures. This study analyzes the functional relevance of certain polymorphisms of the SOST promoter region, and their relationship with the expression and methylation of this gene
Material and method:
Alleles of the rs851054, rs851056, rs10534024, rs1234612 polymorphisms and DNA methylation were analyzed by pyrosequencing in serum and bone samples of 33 patients undergoing hip replacement. In addition, SOST expression was studied in bone samples. Also, different alleles of the SOST promoter were cloned into double reporter vectors with the luciferase gene under this promoter and the alkaline phosphatase gene under a constitutive promoter
Results:
Methylation analysis of the SOST promoter region in serum free DNA and bone DNA revealed no statistically significant differences across the alleles of the analyzed polymorphisms (p>0.05). However, transfections with reporter vectors showed high transcriptional activity, regardless of the vector used.
Conclusions:
We have not found a clear association between the different alleles and the DNA methylation of the SOST promoter region. Further studies are needed to determine the polymorphisms' functional effects on the methylation and expression of the SOST gene and the consequences on bone mass.
Key words: serum free DNA; PMMA; DNA methylation; polymorphisms; sclerostin; osteoporosis; gene regulation
INTRODUCTION
Genome-wide association studies (GWAS) and candidate gene studies have found some single nucleotide polymorphisms (SNPs) in the SOST gene, which encodes sclerostin, associated with bone mineral density (BMD) and predisposition to fractures1 2 3-4. However, the mechanism responsible for this association is unknown. Among the general mechanisms by which genetic variants predispose to complex diseases are epigenetic mechanisms, such as DNA methylation, that modulate gene transcription directly (locally) or indirectly (remotely)5. In this sense, it should be noted that the DNA methylation of the SOST promoter is inversely related to the gene expression levels of the gene6.
DNA methylation is an epigenetic mark that consists in the addition of a methyl group at the 5 'position of the cytosine ring, usually in cytosines that precede guanine, forming the so-called CpG sites. They are distributed throughout the genome and abundant in some specific regions, such as promoters, called CpG islands. Methylation levels of CpG sites and/or islands have specific profiles according to the tissue of origin and modulate gene expression in many tissues, including bone7 8 9-10.
Circulating cell free DNA (cfDNA) is present in fluids, such as urine, synovial fluid, plasma or serum, and it is an interesting molecular biomarker because it is easy to obtain without using invasive procedures11. cfDNA is being extensively studied as a biomarker in the oncology field, for the amount of cfDNA increases with the presence of several tumors. In addition, tumors accumulate specific mutations, which allow them to be differentiated from other DNA sequences with different origin12,13.
Therefore, cfDNA is a promising marker for the detection, diagnosis, prognosis, monitoring and treatment of various diseases14.
Previously, we have demonstrated specific DNA methylation patterns in osteoblast and mesenchymal stem cells (hMSCs) derived from osteoporotic patients. These differentially methylated regions are enriched in genes associated with cell differentiation and skeletal development, in hMSCs15and osteoblasts16, respectively. Specifically, we have previously verified that DNA methylation levels of the SOST promoter regulate gene expression in osteoblasts. Hence, DNA demethylation induces SOST expression, even in cells in which this gene is normally repressed17,18.
This study aims to determine DNA methylation levels in the SOST promoter from serum free DNA and the possible relationship with some polymorphisms previously associated with BMD. In addition, we considered the effects of these polymorphisms on the expression of sclerostin.
MATERIAL AND METHODS
Patient selection
Femoral heads were obtained from 33 patients undergoing hip replacement surgery for osteoporotic fracture (FRX; n=15) or osteoarthritis (OA; n=18). Patients with secondary osteoporosis, secondary osteoarthritis or fractures due to high-energy trauma were excluded. Patients' age ranged between 61 and 91 years. From each patient, samples of bone tissue, blood and serum were obtained. The serum was used to isolate free DNA and study sclerostin promoter methylation. Blood samples were used to obtain genomic DNA in order to analyze the polymorphisms of interest.
The study was approved by the Ethics Committee in Clinical Research of Cantabria and patients gave their written informed consent.
DNA isolation
Trabecular bone samples from the central part of the femoral heads were obtained with a trocar. They were instantly frozen with liquid nitrogen and homogenized with a polytron in lysis buffer and proteinase K. After an overnight incubation at 55ºC, DNA was extracted with phenol: chloroform: isoamyl alcohol, as previously published16. Cell free serum DNA was extracted from two 1 ml aliquots of serum, proccessed in parallel (2 ml of serum per patient for analysis). To each aliquot, in a 15 ml falcon tube, we added 500 µl of lysis buffer (Tris-HCl, EDTA, sodium acetate and SDS) and 5 µl of proteinase K (20 mg/ml). The mixture was incubated for 1 hour in a water bath at 56ºC. DNA was extracted, as with bone, by using phenol: chloroform: isoamyl alcohol. The pellet (not visible) was allowed to dry at room temperature and resuspended with 20 µl of distilled water. Blood cell DNA was extracted with the Illustra blood genomic kit Prep Mini Spin (GE Healthcare Life Sciences, Marlborough, USA.).
Genotyping
Blood cell DNA was quantified by the Qubit procedure (Thermofisher Scientific, Waltham, USA). Four SOST SNPs previously associated with bone mineral density (rs851054, rs851056, rs1234612 and rs10534024) were analyzed by using assays with Taqman probes (Thermofisher).
DNA methylation analysis
500 ng of bone DNA per sample was used to modify with bisulfite with the EZ DNA Methylation-Gold methylation kit (ZymoResearch, Irvine, USA), following the manufacturer's instructions. On the other hand, the whole amount of DNA isolated from serum (20 µl of the resuspended DNA) was used and also subjected to bisulfite modification with the EZ DNA Methylation-Gold kit. The level of CpG methylation in the region of the SOST promoter was analyzed by pyrosequencing (PyromarkQ24 Advanced System®). The primers used for PCR amplification and sequencing were designed with the PyroMark assay designer (Qiagen NV, Hilden, Germany) (Sense primer. 5'-TGGTGGGGTGATAAATGAATT-3'; Antisense primer. 5'-TGGTGGGGTGATAAATGAATT-3'; Sequencing primer 5'-ATTTGGTTTGAGAAATGG-3 '). The PCR was carried out with a biotinylated primer, which allows its purification in a single-stranded DNA template, using the PyromarkQ24 vacuum workstation (Qiagen N.V., Hilden, Germany) (according to the manufacturer's instructions). Finally, pyrosequencing reactions and methylation quantification were carried out in a PyroMark Q24 Advance System (Qiagen N.V., Hilden, Germany).
The region where methylation was studied was located near the polymorphisms examined, approximately 300 base pairs upstream the transcription start site (Figure 1A).

Figure 1. Diagram of the promoter region of the SOST gene and location of the analyzed polymorphisms, with the distance to the transcription start point (TSS). The CpG dinucleotide chosen to determine the methylation levels of is also shown. (B) Reporter vectors with the luciferase gene (G-LUC) under SOST promoter and alkaline phosphatase (SEAP) under the constitutive cytomegalovirus (CMV) promoter. Two vectors were used, with a different haplotype of the frequent polymorphisms of the region (rs851054, rs851056 and rs851057)
SOST expression and sclerostin levels
Serum sclerostin levels were analyzed by ELISA (Teco Medical Group, Sissach, Switzerland). The sensitivity of this kit is 0.05-3 ng/ml.
SOST expression in bone was analyzed by quantitative real-time PCR (RT-qPCR). RNA was extracted from frozen bone biopsies homogenizing with trizol, isolating with chloroform and precipitating the RNA with isopropanol. Complementary DNA (cDNA) was synthesized with the TaKaRa PrimeScript RT kit (TaKaRa, Shiga, Japan). We used 1 µg of starting RNA, random hexamers and oligo-dT as primers, with the protocol recommended by the manufacturer. The transcript levels were evaluated by RT-qPCR using commercially available Taqman assays (Thermofisher Scientific) in an Applied Biosystems 7300 real-time PCR system. We used GAPDH and TBP as reference genes.
Reporters vectors and analysis of transcriptional activity
The SOST promoter reporter vector (HPRM50859-PG04; GeneCopoeia, Rockville, USA) was acquired. In addition, a second vector was obtained with the same sequence, but varying the haplotype (rs851054 G/A; rs851056 C/G; rs851057 C/G). Both vectors have the luciferase gene under the SOST promoter sequence and the bioluminescent alkaline phosphatase gene under a constitutive promoter (Figure 1B). This dual vector allows to normalize the signal and objectively quantify the signal generated by each transfected promoter. Likewise, we obtained a vector with an empty promoter (pEZX-PG04; GeneCopoeia, Rockville, USA) on the luciferase sequence, as a negative control for transfection.
Transfection of the different vectors was carried out with Lipofectamine 3000 (Thermofisher Scientific, Waltham, USA). For the transfection experiments 50,000 cells (HEK-293T) were seeded per well, in a 24-well plate, by triplicate. The next day, with a confluence of 80%, approximately, 500 ng of each vector was transfected, in independent wells, using Lipofectamine 3000 according to the manufacturer's recommendations. The luciferase and alkaline phosphatase signal was analyzed after 24 h, 48 h and 72 h, by using the Secrete-Pair Dual Luminescence Assay Kit (GeneCopoeia, Rockville, USA).
Analysis of the results
The presence of linkage imbalance and haplotypic distribution was analyzed with the Haploview program19.
Statistical analyzes for this study were carried out using version 3.6.0 of the R software. Alleles were compared with respect to their level of SOST promoter methylation and/or SOST expression in bone by analysis of variance (ANOVA). The comparison between patient groups was performed using Student's t. In all cases, p values less than 0.05 were considered as significant.
RESULTS
Serum free DNA methylation samples were analyzed, in duplicate, by pyrosequencing. DNA methylation levels were taken as reliable when the signal strength was solid. Variability among serum duplicates was small, with an average standard error of 3.89%.
Serum free DNA methylation analysis did not reveal statistically significant differences in relation to the various alleles of the analyzed polymorphisms (rs851054, rs851056, rs1234612 and rs10534024) (Figure 2). We did not find differences in bone DNA methylation in association with the aforementioned polymorphisms (Figure 3).

Figure 2. Methylation level of the SOST promoter in serum cell-free DNA ofindividuals (n=33) genotyped for each of the 4 polymorphisms (rs851054, rs851056, rs10534024 and rs1234612). P-values of the analysis of the variance of methylation levels across genotypes

Figure 3. Methylation level of the SOST promoter in bone DNA of individuals (n=33) genotyped for each of the 4 polymorphisms (rs851054, rs851056, rs10534024 and rs1234612). P-values of the analysis of the variance of methylation levels across genotypes
In addition RNA was also obtained from bone biopsies in order to study the endogenous expression of SOST in bone. The results obtained by RT-qPCR did not reveal statistically significant differences in the endogenous expression of SOST in bone, in relation to the polymorphisms analyzed (Figure 4).

Figure 4. SOST expression in bone. Samples of individuals (n=33) genotyped for each of the 4 polymorphisms (rs851054, rs851056, rs10534024 and rs1234612). Expression levels were calculated by RT-qPCR, standardized by the reference genes (GAPDH and TBP) and are expressed as deltaCt. P-values of the analysis of the variance of methylation levels across genotypes
It should be noted that 3 of the 4 SNPs were in strong linkage imbalance, with D 'of 1 and close correlation between their alleles (R2of 0.83-1). The other polymorphism, rs1234612, was not part of that block (Figure 5). Combined haplotype or genotype analysis did not reveal statistically significant associations with methylation or gene expression (data not shown). Transfections with reporter vectors, which carried the promoter sequence of the SOST gene, showed high transcriptional activity, regardless of the alleles present in the vector. In fact, it increased up to 20 times at 24 hours with respect to the empty vector. However, both constructions, with opposite alleles, showed a similar activity (Figure 6).

Figure 5. Linkage disequilibrium between the polymorphisms analyzed. The numbersrepresent the distance values D', which can range from 0 to 1. The figure represents the values multiplied by 100. In the case of color boxes, the value is 1
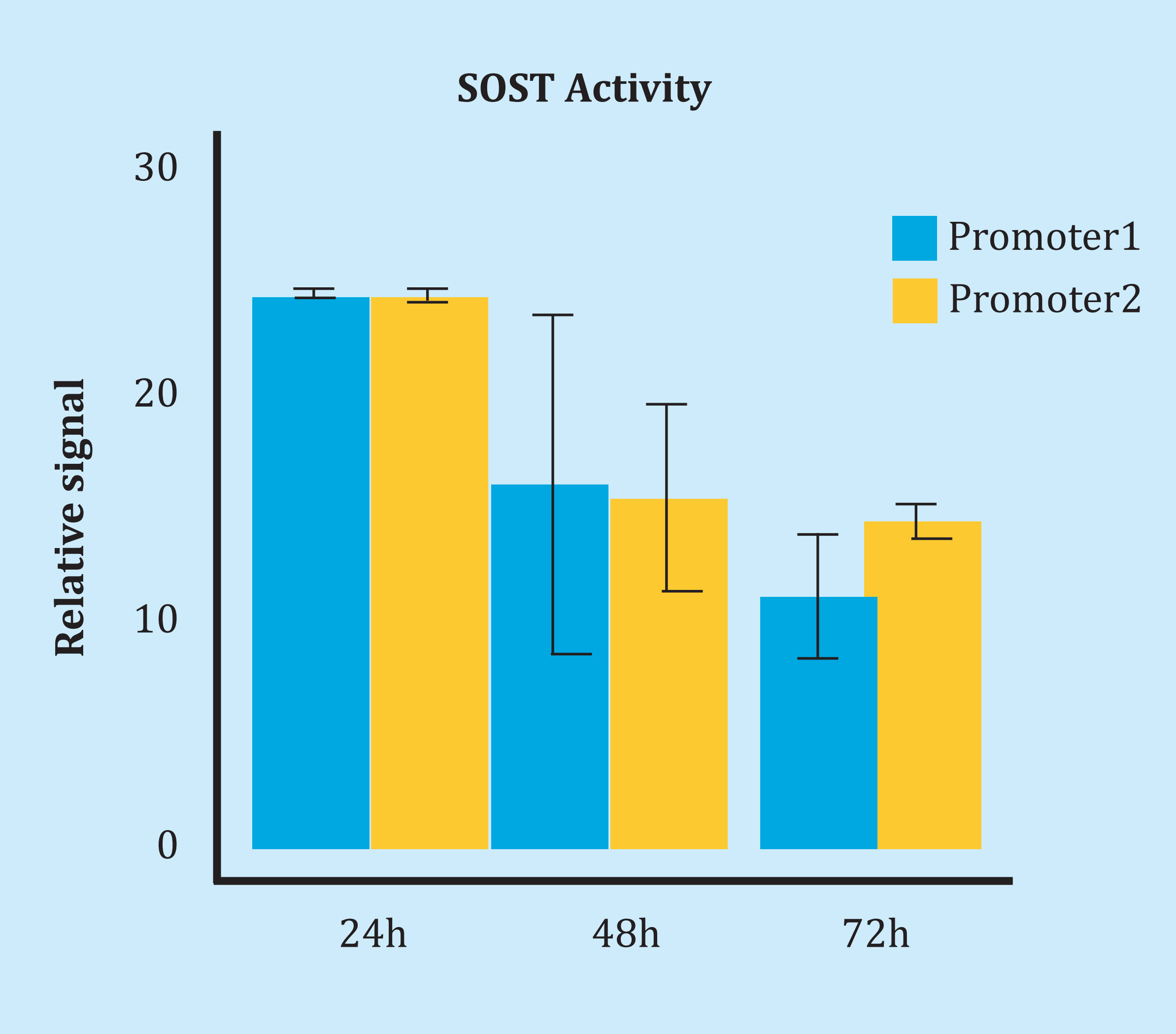
Figure 6. SOST promoter activity measurement by transfecting vectors with different SOST sequences (promoter 1 and promoter 2). Each reporter has the oppositehaplotype for polymorphisms rs851054, rs851056 and rs851057. The relative signal was calculated as the ratio of luciferase activity and the alkaline phosphatase activity ratio (SEAP). Subsequently it was compared with the ratio observed after transfecting an empty vector (this is, without the SOST promoter, but with SEAP activity). Bars show the standard error
DISCUSSION
Sclerostin is a potent inhibitor of the Wnt pathway. It blocks the co-receptors Lrp4, 5 and 6, thus preventing the receptor activation by Wnt ligands. Sclerostin has an important role in bone biology. Mice with SOST gene deletion have ibcreased bone formation and bone mass20. In contrast, overexpression of SOST in osteoblasts decreases bone mass21. In addition, certain SOST gene mutations that cause a loss of sclerostin in humans are associated with high bone formation activity and high BMD, causing Van Buchem disease or sclerosteosis22,23. Conversely, a monoclonal antibody that blocks sclerostin (romosozumab) has recently been approved by the FDA (US Food and Drug Administration) for osteoporosis treatment, after observing that it increased bone mass in animal studies and in humans24,25.
Several studies suggest that some allelic variants of the SOST gene may influence BMD and the risk of osteoporosis26,27. Since these are noncoding variants, presumably they influence the expression of SOST gene. On the other hand, we have previously been able to demonstrate the importance of DNA methylation in the regulation of sclerostin expression in the osteoblastic lineage18. Likewise, in several studies it has been shown that genetic variants can influence DNA methylation and SOST gene expression5. Hence, the objective of this study was to explore the functional impact of some frequent polymorphisms in the promoter region of the SOST gene, specifically, their effect on methylation and gene expression. However, despite its association with BMD15, we have not found any significant association between these allelic variants and DNA
methylation levels, nor between allelic variants and gene expression levels, whether the analysis was performed at the single SNP level, or at the combined genotype or haplotype levels. In line with this, transfection experiments with reporter vectors have not revealed differences between allelic variants of the promoter region and transcriptional activity. Therefore, our study does not support the existence of an influence of these polymorphisms on the expression of the sclerostin gene, neither direct nor mediated through changes in promoter methylation.
There are several limitations that must be considered when interpreting these negative results. First, the study of association between allelic variables and DNA methylation is limited to a specific region of the promoter. A study of other CpG sites along the SOST region would be needed to obtain a more comprehensive understanding of the potential influence of DNA polymorphisms on DNA methylation. Second, the effect of the polymorphisms studied may depend on other polymorphisms in linkage disequilibrium. In addition, those polymorphisms may be in regions far from the promoter, such as regulatory regions (enhancer) or even in other chromosomes. This fact also limits the analysis with reporter vectors, in which only the promoter region of SOST is found. Finally, reporter vectors are transfected in vitro, and their DNA sequences are demethylated, so that the in vivosituation is not properly recapitulated. Another limitation of this study is the presence of samples obtained from patients with different diseases (osteoporotic fractures and osteoarthritis), that may influence methylation levels distinctly. However, similar results were obtained in the stratified analysis. Finally, the sample size determines the ability to demonstrate subtle differences between polymorphisms, especially in the analysis of combined polymorphisms. In any case, it is important to note that these results do not question the importance of sclerostin in regulating bone cell activity, which has been demonstrated in numerous experimental and clinical studies.
In conclusion, we have not seen a clear association between the different alleles and the DNA methylation of the promoter region of the SOST gene. Therefore, the association of these polymorphisms with BMD does not appear to be due to direct influences on the promoter activity, or to changes in promoter methylation. It can be assumed, therefore, that it is mediated by complex interactions that take place with distant regions of the chromatin. On the other hand, this study raises the possibility of using serum free DNA as a biomarker in some skeletal disorders.
REFERENCES
1 Lhaneche L, Hald JD, Domingues A, Hannouche D, Delepine M, Zelenika D, et al. Variations of SOST mRNA expression in human bone are associated with DNA polymorphism and DNA methylation in the SOST gene. Bone. 2016;92:107-15. [ Links ]
2 Zhou P, Xu X, Zhang Z, Liao E, Chen D, Liu J, et al. SOST polymorphisms and response to alendronate treatment in postmenopausal Chinese women with osteoporosis. Pharmacogenomics. 2015; 16(10):1077-88. [ Links ]
3 Ye W, Wang Y, Mei B, Hou S, Liu X, Wu G, et al. Computational and functional characterization of four SNPs in the SOST locus associated with osteoporosis. Bone. 2018;108:132-44. [ Links ]
4 Kuipers AL, Zhang Y, Yu S, Kammerer CM, Nestlerode CS, Chu Y, et al. Relative influence of heritability, environment and genetics on serum sclerostin. Osteoporos Int. 2014;25(3):905-12. [ Links ]
5 Wang H, Lou D, Wang Z. Crosstalk of genetic variants, allele-specific DNA methylation, and environmental factors for complex disease risk. Front Genet. 2018;9:695. [ Links ]
6 Delgado-Calle J, Sañudo C, Bolado A, Fernández AF, Arozamena J, Pascual-Carra MA, et al. DNA methylation contributes to the regulation of sclerostin expression in human osteocytes. J Bone Miner Res. 2012;27(4):926-37. [ Links ]
7 Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. Schubeler D, editor. PLoS Genet. 2009;5 (8):el000602. [ Links ]
8 Ghayor C, Weber F. Epigenetic regulation of bone remodeling and its impacts in osteoporosis. Int J Mol Sci. 2016;17(9):1446. [ Links ]
9 Wang P, Cao Y, Zhan D, Wang D, Wang B, Liu Y, et al. Influence of DNA methylation on the expression of OPG/ RANKL in primary osteoporosis. Int J Med Sci. 2018;15(13):1480-5. [ Links ]
10 Reppe S, Lien TG, Hsu Y-H, Gautvik VT, Olstad OK, Yu R, et al. Distinct DNA methylation profiles in bone and blood of osteoporotic and healthy postmenopausal women. Epigenetics. 2017;12(8):674-87. [ Links ]
11 Nagy B. Cell-free nucleic acids in prenatal diagnosis and pregnancy-associated diseases. EJIFCC. 2019;30(2): 215-23. [ Links ]
12 Yang X, Zhang K, Zhang C, Peng R, Sun C. Accuracy of analysis of cfDNA for detection of single nucleotide variants and copy number variants in breast cancer. BMC Cancer. 2019;19(1):465. [ Links ]
13 Herrmann S, Zhan T, Betge J, Rauscher B, Belle S, Gutting T, et al. Detection of mutational patterns in cell free DNA (cfDNA) of colorectal cancer by custom amplicon sequencing. Mol Oncol. 2019; 13 (8): 1669-83. [ Links ]
14 Pös O, Biró O, Szemes T, Nagy B. Circulating cell-free nucleic acids: characteristics and applications. Eur J Hum Genet. 2018;26(7):937-45. [ Links ]
15 del Real A, Pérez-Campo FM, Fernández AF, Sañudo C, Ibarbia CG, Pérez-Núñez MI, et al. Differential analysis of genomewide methylation and gene expression in mesenchymal stem cells of patients with fractures and osteoarthritis. Epigenetics. 2017;12(2):113-22. [ Links ]
16 Delgado-Calle J, Fernández AF, Sainz J, Zarrabeitia MT, Sañudo C, García-Renedo R, et al. Genome-wide profiling of bone reveals differentially methylated regions in osteoporosis and osteoarthritis. Arthritis Rheum. 2013;65(l):197-205. [ Links ]
17 Delgado-Calle J, Pérez-Campo FM, Riancho JA. Avances en el estudio de los mecanismos involucrados en la modulación de la expresión de esclerostina en células humanas. Rev Osteoporos Metab Miner. 2014;6(4):103-8. [ Links ]
18 Delgado-Calle J, Sañudo C, Bolado A, Fernández AF, Arozamena J, Pascual-Carra MA, et al. DNA methylation contributes to the regulation of sclerostin expression in human osteocytes. J Bone Miner Res. 2012;27(4):926-37. [ Links ]
19 Barrett JC, Fry B, Mailer JDM. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263-5. [ Links ]
20 Sebastián A, Loots GG. Genetics of Sost/SOST in sclerosteosis and van Buchem disease animal models. Metabolism. 2018;80:38-47. [ Links ]
21 Pérez-Campo FM, Santurtún A, García-Ibarbia C, Pascual MA, Valero C, Garcés C, et al. Osterix and RUNX2 are Transcriptional Regulators of Sclerostin in Human Bone. Calcif Tissue Int. 2016; 99(3):302-9. [ Links ]
22 He W, Chen C, Pan C, Zhang M, Yu X, Wang D, et al. Sclerosteosis caused by a novel nonsense mutation of SOST in a consanguineous family. Clin Genet. 2016;89(2):205-9. [ Links ]
23 Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, et al. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15 (7):928-35. [ Links ]
24 Cosman F, Crittenden DB, Adachi JD, Binkley N, Czerwinski E, Ferrari S, et al. Romosozumab Treatment in Pöstmenopausal Women with Osteoporosis. N Engl J Med. 2016;375(16):1532-43. [ Links ]
25 McClung MR. Romosozumab for the treatment of osteoporosis. Osteoporos Sarcopenia. 2018;4(l):ll-5. [ Links ]
26 Uitterlinden AG, Arp PP, Paeper BW, Charmley P, Proll S, Rivadeneira F, et al. Polymorphisms in the sclerosteo-sis/van Buchem disease gene (SOST) region are associated with bone-mineral density in elderly whites. Am J Hum Genet. 2004;75(6):1032-45. [ Links ]
27 Lhaneche L, Hald JD, Domingues A, Hannouche D, Delepine M, Zelenika D, et al. Variations of SOST mRNA expression in human bone are associated with DNA polymorphism and DNA methylation in the SOST gene. Bone. 2016;92:107-15. [ Links ]
Received: July 10, 2019; Accepted: November 16, 2019











 texto em
texto em 


