Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Revista de Osteoporosis y Metabolismo Mineral
versión On-line ISSN 2173-2345versión impresa ISSN 1889-836X
Rev Osteoporos Metab Miner vol.12 no.2 Madrid abr./jun. 2020 Epub 05-Oct-2020
https://dx.doi.org/10.4321/s1889-836x2020000200002
ORIGINALS
A sensitive method for monitoring the migration of mesenchymal stem cells from bone marrow in murine models
1Internal Medicine Service. Marqués de Valdecilla University Hospital. University of Cantabria. Valdecilla Research Institute (IDIVAL). Santander (Spain).
2Traumatology Service. Marqués de Valdecilla University Hospital. University of Cantabria. Valdecilla Research Institute (IDIVAL). Santander (Spain).
3Maxillofacial Surgery Service. Marqués de Valdecilla University Hospital. Santander (Spain).
4Legal Medicine Unit. University of Cantabria. Valdecilla Research Institute (IDIVAL). Santander (Spain).
Objetive:
Mesenchymal stem cells (MSCs) are commonly used in regenerative therapy of human diseases. In murine models, in which human MSCs are transplanted, distinguishing the origin of the identified MSCs in the organs of mice is important. The objective of this study was to determine the performance of PCRbased analysis of human Alu sequences to detect human DNA after infusion of human bone marrow stem cells (hBMSCs) in immunodeficient mice.
Material and method:
HBMSCs were obtained from the femoral head of patients undergoing hip replacement surgery. 106 hBMSCs were infused intravenously by injection into the retroorbital sinus of NOD/SCID mice. The presence of human DNA in lung, liver and bone was then assessed.
Results:
In in vitro DNA mixtures, human DNA was easily detected with a good logarithmiclinear relationship. Similarly, when human and mouse osteoblasts were mixed, 110 cells were easily detected among 105 mouse cells. Likewise, human DNA was detected in the lungs 1 and 7 days after cell infusions in NOD/SCID mice. However, human DNA was inconsistently detected in the liver and bones.
Conclusion:
Detecting Alu sequences is an effective procedure to observe human DNA. The results confirm that most intravenously injected hBMSCs are trapped in the lungs. Thus, for the treatment of skeletal disorders, procedures are needed to increase the migration of these cells to the bone.
Key words: Mesenchymal stem cells; Osteoporosis; Cell migration; Regenerative therapy; Alu sequences
INTRODUCTION
Osteoporosis is the most frequent bone disease, characterized by low bone mass and alteration of the microstructure. This is due to an imbalance between bone formation and bone resorption that causes loss of connections among the different bone trabeculae, a greater thinning and cortical bone porosity. Consequently, there is greater bone fragility and an increased risk of fractures (Fx)1,2.
Osteoblasts, cells specialized in bone formation, originate from the differentiation of mesenchymal stem cells (MSCs)3. These cells are multipotent and can differentiate into a wide variety of mesoderm cell types, such as osteoblasts, adipocytes, or chondrocytes. MSCs are highly interesting candidates for regenerative medicine, because they migrate to skeletal lesions where they have the capacity to form new bone4. The many relevant published studies show the importance of MSCs in tissue engineering and regenerative medicine5,6. In addition, there are currently more than 250 clinical trials with MSCs, as reflected in the clinical trial database (clinicaltrials.gov).
Imaging techniques such as magnetic resonance imaging and positron emission tomography, and cells previously labeled with a fluorophore are used to monitor transplanted human cells in animal models to detect the signal in vivo7,8. An alternative approach is to detect the presence of human DNA in ex-vivo animal models. So, once the treatment is complete, the presence of DNA of human origin is accessed in the target organ by realtime quantitative PCR (qPCR)9-11. Alu sequences or elements are short, repetitive, intercalating elements of the genome (SINE), approximately 300 base pairs in length. There are more than 1 million copies of Alu sequences in the human genome, occupying about 10% of the entire genome12,13. Given their small size, specific distribution among species and high number of copies, they are a very useful target for detecting human cells. However, most of the Alubased experimental techniques to detect only human genomic DNA do not reach the limits of sensitivity and specificity necessary to distinguish them from DNA from other primates or rodents13,14. Funakoshi et al. have developed a highly sensitive and specific Alubased quantitative realtime PCR method to discriminate human cells from rodent cells, to avoid possible crossreactions11.
The objective of this study was to determine the performance of PCRbased analysis of human Alu sequences to detect human DNA after infusion of human bone marrow stem cells (hBMSCs) in immunodeficient mice (NOD/SCID).
MATERIAL AND METHODS
Isolation of hBMSCs
HBMSCs were obtained from the femoral head of patients undergoing hip replacement surgery. The study was approved by the Cantabria Clinical Research Ethics Committee and patients gave their written informed consent. Cylinders of trabecular bone were removed from the femoral head with a trocar and these were washed in PBS to obtain the bone marrow cells. Ficoll gradients were centrifuged to obtain the mononuclear layer, the one that was finally cultivated to attain an 80% state of confluence.
NOD/SCID mice and cell infusion
NOD/SCID immunodeficient mice, obtained from Charles River Laboratories International, Inc. (Wilmington, Massachusetts, USA), were injected with 106 hBMSCs intravenously infused into the retroorbital sinus.
DNA isolation and real-time quantitative PCR
The mouse femur and human bone cylinders were homogenized with a polytron in lysis buffer and proteinase k, which was stored in an overnight incubation at 55°C with shaking. The soft tissues, lung and liver, were directly homogenized in lysis buffer and proteinase k. The DNA was then isolated with phenol: chloroform: isoamyl alcohol, and precipitated with 100% ethanol. The presence of human DNA in the DNA extracted from these organs (lung, liver and bone) was evaluated by realtime PCR, with a hybridization temperature of 56ºC for 40 cycles, using the primers and protocol proposed by Funakoshi11 (Table 1).
Negative controls without DNA (NTC) and DNA extracted from mouse tissues without hBMSCs were included in all cases. Likewise, DNA extracted from artificial mixtures of human and mouse cells, as well as mixtures of purified human DNA and murine DNA, were analyzed. The threshold cycle (Ct) of each sample, that is, the amplification cycle from which the amplicons were detectable, was estimated. Logically, there is an inverse relationship between the amount of target DNA present in the sample and the Ct.
Written informed consent was obtained from patients who donated hBMSCs, in accordance with procedures approved by the Cantabria Clinical Research Ethics Committee. Regarding animal experiments, the protocol was approved by the Research Ethics Committee of the University of Cantabria and the Ministry of Health of Cantabria, as established by current regulations.
RESULTS
Mixtures of human and mouse DNA
In the methods of detecting human DNA in a different organism, such as the mouse, high technical sensitivity and specificity are critical. For this, the first evaluation of the detection technique used in this article was carried out with DNA mixtures and with mixtures of different numbers of cells of human and mouse origin. A spectrophotometer (DeNovix DS11, Wilmington, USA) was used to assess the amount of DNA in each sample. First, 100 ng/µL human DNA standard solutions were mixed with 100 ng/µL mouse DNA in a 1:1 ratio and up to 8 serial dilutions 1:10 were made in mouse DNA. Thus, progressive dilutions of human DNA obtained in the presence of mouse DNA practically constant. The expression levels on the Ct scale were 11.5; 14.6; 17.2; 21.0; 23.8; 26.8; 30.0; 32.5 and 33.6; for 100 ng/µL, 10 ng/µL, 1 ng/µL, 0.1 ng/µL, 0.01 ng/µL, 1 pg/µL, 0.1 pg/µL, 0.01 pg/µL and 0.001 pg/µL of human DNA, respectively (r2=0.992; p<0.0001) (Figure 1). In several independent experiments, the threshold cycle (Ct) for NTC was 34.6±1.8, so 31 (2 standard deviations below the NTC mean) were considered as the maximum Ct to consider positive. the presence of human DNA in a sample. No signal was detected when up to 100 ng of mouseonly DNA was analyzed (Ct was 34.7±1.6).

Figure 1. A) Detection of different concentrations of human DNA in a high concentration of mouse DNA. B) The same results using a logarithmic scale on the axis of abscissa indicate the expected log-linear relationship. The figure indicates the r2 value of the Pearson regression and the significant p value for the linear regression. The broken line in A) and B) shows the threshold value of detection of DNA of human origin when mixing different concentrations
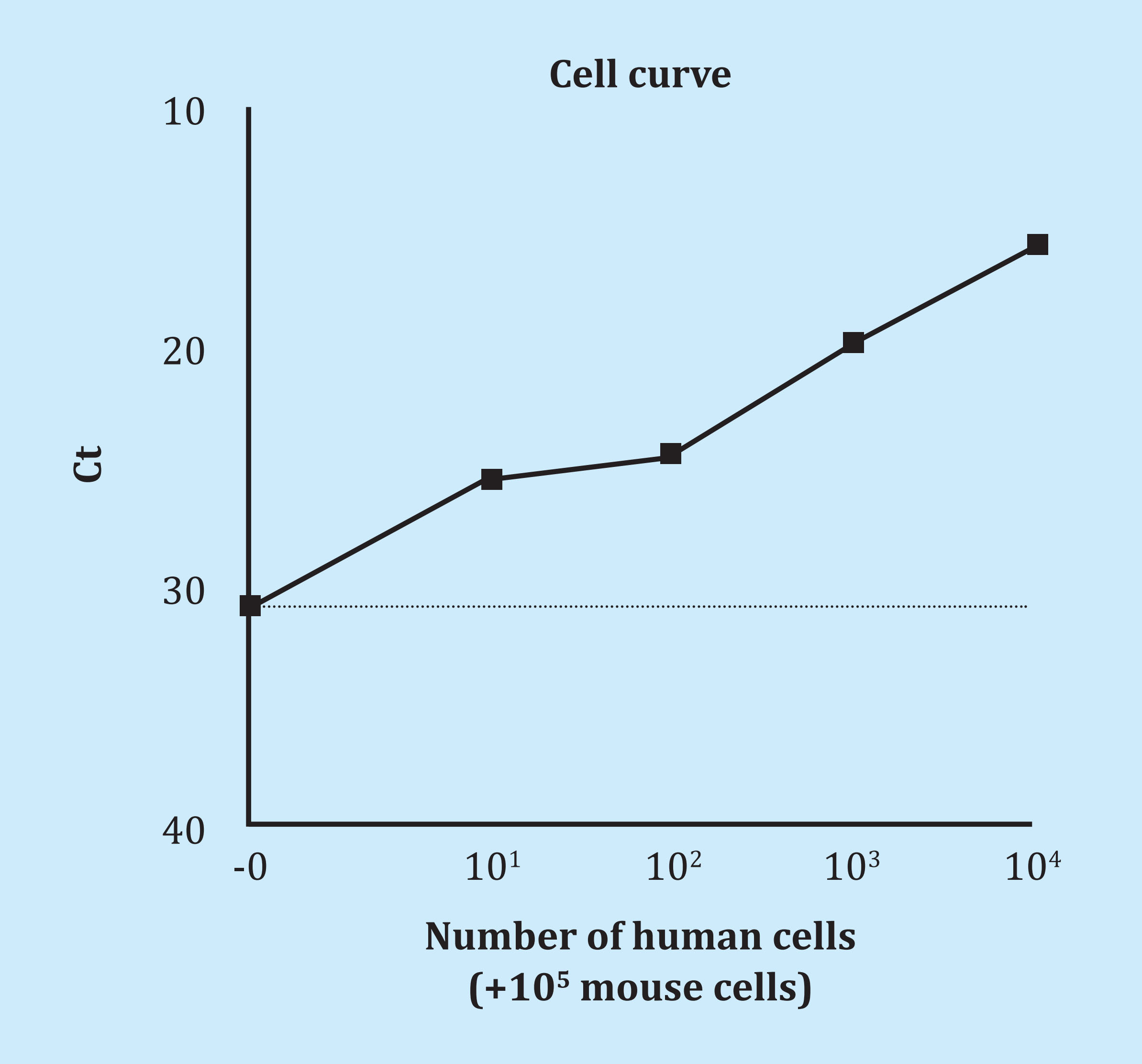
Subsequently, human and mouse osteoblasts were mixed in different proportions prior to DNA extraction to simulate the technique in a real way. The 10 human cells were easily detected in a mixture of 105 mouse cells (Ct 25.6) and even a single cell was close to the detection limit (Ct 30.8) (Figure 2).
Analysis of human DNA in mouse tissues
Tissues from nonfractured NOD/SCID mice treated with intravenous hBMSCs were then analyzed. These levels were also compared with those detected in diluted human bone DNA samples, as positive controls. Human DNA could be located in the lungs on the first day and 7 days after cell infusions (Ct 22.6±0.7 and 30.6±3.7, respectively). However, human DNA was inconsistently detected in the liver and bones (Figure 3). There is a decrease in human DNA between days 1 and 7 in the lung samples, but these differences are not significant. However, when comparing day 1 human DNA levels with samples without DNA they are significant (Figure 4).

Figure 3. Detection of human Alu sequences in different mouse tissues 1 and 7 days after infusion of BMSCs intravenously. In orange are the control samples to which no cells were injected. The broken line shows the threshold value for detection of DNA of human origin. (●) Liver; (■) Lung; (▲) Bone. Day 1 geometric figures show the mean of 3 mice; those of day 7 of 4 mice; and those without cells from 2 mice. The downward triangles (▼), green in color, are samples of bone of human origin
DISCUSSION
Reparing bone fractures is a complex process, where there are a series of molecular mechanisms regulated by various factors that lead to new bone formation. This repair can sometimes be altered by aging and by different bone disorders, such as osteoporosis or avascular necrosis, among others5. Regenerative therapy attempts to solve these imbalances by avoiding allogeneic transplant rejection and adverse immune reactions. For this, new osteoinductive biomaterials, osteogenic regulation factors and MSCs15 have been used. These are of great interest and numerous studies involving MSCs have been published. MSCs are characterized by having a high capacity for renewal and also being able to form new cell types of mesodermal origin, such as osteoblasts or adipocytes. Furthermore, they have immunomodulatory effects and secrete factors that induce cell differentiation5,16.
The physiological function and repair capacity of human MSCs are commonly studied in xenografts carried out in rodents. After intravenous xenotransplantation, cells can circulate widely throughout the body and their tropism by different organs needs to be studied. Consequently, it is necessary to study these cells' distribution after blood infusion. To do so, a highly sensitive, specific method of detecting small populations of human cells among the cells of the recipient organism is needed. Funakoshi et al. have developed a qPCR system, theoretically very sensitive and specific, that allows us to detect these small populations of human MSCs that have survived after their infusion in mice. The mechanism, based on Alu sequences that differ from each other in terms of species evolution and can specifically detect those of uniquely human origin11. Due to the extremely high number of copies of the Alu sequence in the human genome, a single primer could amplify the inter-Alu genomic sequence, which can result in the formation of amplified products with unpredictable and complex patterns. To minimize the effects of such non-specific signals, the method uses hydrolysis probes, which hybridize to the sequence to be amplified between both primers. Still, in this reaction there are nonspecific hybridizations to the mouse genome that cause an unavoidable background fluorescence signal, which is considered technical noise. Our objective was to confirm the usefulness of this methodology in our model.
In fact, with this procedure we were able to detect very low concentrations of human DNA among a high concentration of mouse DNA, specifically up to 0.01 pg/µL of human DNA between 100 ng/µL of mouse. In cell mixtures, the detection threshold was 1-10 human cells in 105 mouse cells.
HBMSCs were injected intravenously into mice. This procedure verified that, after the first 24 hours and the seventh day, they were only detectable in the lung (they were not consistently detected in liver or bone). Various strategies are being tested to increase the tropism of hBMSCs to bone tissue. One of them is based on modifying membrane proteins, with specific glycosylation particles that allow extravasation and a greater tropism for bone17.
In conclusion, the results confirm that the majority of hBMSCs injected intravenously into NOD/SCID mice are trapped in the lungs and are rapidly lost. Therefore, procedures are needed to increase the tropism of these cells to bone if hBMSCs areto be used in systemic regenerative skeletal procedures.
REFERENCES
1 Eastell R, O'Neill TW, Hofbauer LC, Langdahl B, Reid IR, Gold DT, et al. Postmenopausal osteoporosis. Nat Rev Dis Prim. 2016;2(1):16069. [ Links ]
2 Díaz Curiel M. Osteoporosis: concepto. Fisiopatología. Clínica. Epidemiología. Rev Osteoporos Metab Miner. 2018;10 (1 Suplemento):2-4. [ Links ]
3 Katsimbri P. The biology of normal bone remodelling. Eur J Cancer Care (Engl). 2017;26(6). [ Links ]
4 Han Y, Li X, Zhang Y, Han Y, Chang F, Ding J. Mesenchymal stem cells for regenerative medicine. Cells. 2019;8(8):886. [ Links ]
5 Iaquinta MR, Mazzoni E, Bononi I, Rotondo JC, Mazziotta C, Montesi M, et al. Adult stem cells for bone regeneration and repair. Front Cell Dev Biol. 2019; 7:268. [ Links ]
6 Brown C, McKee C, Bakshi S, Walker K, Hakman E, Halassy S, et al. Mesenchymal stem cells: Cell therapy and regeneration potential. J Tissue Eng Regen Med. 2019 Sep;13(9):1738-55. [ Links ]
7 Freeman BT, Kouris NA, Ogle BM. Tracking fusion of human mesenchymal stem cells after transplantation to the heart. Stem Cells Transl Med. 2015; 4(6):685-94. [ Links ]
8 Chikate TR, Tang L. Tracking and imaging of transplanted stem cells in animals. Methods Mol Biol. 2019; online ahead of print. [ Links ]
9 Creane M, Howard L, O'Brien T, Coleman CM. Biodistribution and retention of locally administered human mesenchymal stromal cells: Quantitative polymerase chain reaction-based detection of human DNA in murine organs. Cytotherapy. 2017;19 (3):384-94. [ Links ]
10 Schubert R, Sann J, Frueh JT, Ullrich E, Geiger H, Baer PC. Tracking of adipose-derived mesenchymal stromal/stem cells in a model of cisplatin-induced acute kidney injury: Comparison of bioluminescence imaging versus qRT-PCR. Int J Mol Sci. 2018;19(9):E2564. [ Links ]
11 Funakoshi K, Bagheri M, Zhou M, Suzuki R, Abe H, Akashi H. Highly sensitive and specific Alu-based quantification of human cells among rodent cells. Sc iRep. 2017;7(1):13202. [ Links ]
12 Salem A-H, Kilroy GE, Watkins WS, Jorde LB, Batzer MA. Recently integrated Alu elements and human genomic diversity. Mol Biol Evol. 2003;20(8):1349-61. [ Links ]
13 Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3(5):370-9. [ Links ]
14 Jurka J. Evolutionary impact of human Alu repetitive elements. Curr Opin Genet Dev. 2004;14(6):603-8. [ Links ]
15 Iaquinta MR, Mazzoni E, Manfrini M, D'Agostino A, Trevisiol L, Nocini R, et al. Innovative biomaterials for bone regrowth. Int J Mol Sci. 2019;20(3):618. [ Links ]
16 Abdel Meguid E, Ke Y, Ji J, El-Hashash AHK. Stem cells applications in bone and tooth repair and regeneration: New insights, tools, and hopes. J Cell Physiol. 2018;233(3):1825-35. [ Links ]
17 Sackstein R, Merzaban JS, Cain DW, Dagia NM, Spencer JA, Lin CP, et al. Ex vivo glycan engineering of CD44 programs human multipotent mesenchymal stromal cell trafficking to bone. Nat Med. 2008;14(2):181-7. [ Links ]
Received: January 23, 2020; Accepted: April 08, 2020











 texto en
texto en