Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Ars Pharmaceutica (Internet)
versión On-line ISSN 2340-9894
Ars Pharm vol.57 no.3 Granada jul./sep. 2016
https://dx.doi.org/10.30827/ars.v57i3.5329
ORIGINAL ARTICLE
Hospital effluent constitutes a source of vancomycin-resistant enterococci
El efluente hospitalario como fuente de enterococos vancomicina resistentes
Lidia Nuñez1, Carina Tornello1, Noel Puentes1, Elena Espigares2, Elena Moreno2, Miguel Espigares2 and Juan Moretton1
1. Cátedra de Higiene y Sanidad, Faculty of Pharmacy and Biochemistry, University of Buenos Aires, Junín 954/6, Buenos Aires, C1113AAD, Argentina.
2. Department of Preventive Medicine and Public Health, Faculty of Pharmacy, University of Granada, Campus Universitario de Cartuja, Granada, 18071, Spain.
ABSTRACT
Objectives: Enterococci are intrinsically resistant to many commonly used antimicrobial agents. They are able to acquire resistance with relative ease and can spread these genes to other species. Enterococci resistant to antibiotics are associated with the use of these in clinical practice and also the spread of resistant clones in the world. The aim of this work was to compare the characteristics of the strains of vancomycin-resistant enterococci (VRE) isolated from municipal wastewater and hospital effluent.
Methods: Samples were obtained from the effluent of the Hospital Universitario José de San Martín (Buenos Aires) and the municipal wastewater of the city of Buenos Aires.
Results: The bacterial counts of VRE were greater in the hospital effluent, with an odds ratio of 36.4 (95% CI: 26.0-50.8; p<0.0001). The VRE isolated were mainly identified as E. faecium. The results indicate a high prevalence of enterococci resistant to the antibiotics tested.
Conclusion: We may conclude that the effluents of hospitals constitute a source of VRE showing multiple resistance to antibiotics.
Keywords: vancomycin-resistant enterococci; multi-resistance; hospital effluent; antibiotics; municipal wastewater.
RESUMEN
Objetivos: Las especies de enterococos son intrínsecamente resistentes a varios antibióticos, adquieren resistencia con relativa facilidad, y difunden estos genes de resistencia a otras especies. La resistencia a los antibióticos en enterococos está asociada al uso de los mismos en la clínica médica y también a la diseminación de clones resistentes en el mundo. El objetivo de este trabajo fue comparar las características de las cepas de enterococos resistentes a vancomicina (ERV) aisladas en efluentes hospitalarios y aguas residuales urbanas.
Métodos: Se obtuvieron muestras de los efluentes del Hospital Universitario José de San Martín (Buenos Aires) y muestras de aguas residuales urbanas de la ciudad de Buenos Aires.
Resultados: Los recuentos de ERV fueron mayores en los efluentes hospitalarios, siendo la odds ratio 36.4 (IC95%: 26.0-50.8; p<0.0001). Los ERV aislados se identificaron principalmente como E. faecium. Los resultados indicaron una alta prevalencia de enterococos resistentes al resto de los antibióticos ensayados.
Conclusión: Podemos concluir que los efluentes de los centros hospitalarios constituyen una fuente de enterococos de resistencia múltiple a antibióticos.
Palabras clave: enterococos resistentes a vancomicina; multirresistencia; efluente hospitalario; antibióticos; aguas residuales urbanas.
Introduction
Water bodies receive urban wastewater, often inadequately treated, with serious consequences for public health and the environment. Untreated urban sewage contains a high concentration of organic matter and an abundance of pathogenic and saprophytic microorganisms. The water bodies are inherently capable of self-purification, but at times the influx of contaminants exceeds this capacity.
Enterococci, which are part of the natural intestinal flora of animals and humans, can be released to the environment by means of sewage or wastewater.11 Some members of the genus, such as Enterococcus faecalis and Enterococcus faecium, are opportunist pathogens.13 One factor contributing to the pathogenesis of enterococci is their resistance to a broad range of antibiotics. This resistance has increased in recent years.16 Vancomycin is a glycopeptide antibiotic used for serious infections by Gram-positive bacteria when treatment with other antibiotics has failed. The excessive use of this antimicrobial agent has led to the appearance of vancomycin-resistant enterococci (VRE), most notably E. faecalis and E.faecium.4
The epidemiology of infection by enterococci resistant to glycopeptides differs markedly between Europe and the United States. In Europe VRE are frequently isolated from farm animals, associated with a widespread use, until 1997, of avoparcin by bird breeders to promote growth.4 In the United States, avoparcin was never approved for use in breeding animals, for which reason VRE are not found in animals or healthy people. However, nosocomial infection by VRE has undergone a dramatic increase attributed to the generalized use of vancomycin in hospitals.25 In Latin America, the rate of VRE from clinical isolation increased from 5% to 15% between 2003 and 2008; the most significant increase was seen in Brazil, with figures of VRE that increased from 7% to 31% in the same time period.24
Human activities and especially those related with the use of antibiotics when breeding fowl and fish, and in human and veterinary medicine, may be at the root of the high incidence of resistance detected in microorganisms isolated from the environment.2,18 Hospitals represent critical environments for the selection of clinically relevant bacterial resistance, or of genetic determinants, given the amount of antibiotics used and released to the environmental.15 The transmission of enterococci occurs in the hospital environment through contact with objects or the hands of healthcare personnel, whereas in the community the main via of transmission is the consumption of contaminated water or food.5
The objective of this study was to compare the prevalence and the characteristics of the strains of VRE isolated from a hospital effluent and from municipal wastewater.
Materials and methods
Sample collection
Thirty samples of effluent were obtained from the Hospital Universitario José de San Martín (Buenos Aires), at a point before the mixture of their waste with municipal wastewater. This high-level hospital releases approximately 560 m3 per day to the municipal sewage system.21 All the samples of hospital effluent were integrated, through the mixture of four aliquots of three liters (12 liters), between 9 a.m. and 3 p.m., then kept at 4oC until their processing that same day. In parallel, 20 samples of municipal wastewater of the city of Buenos Aires were taken from one of the general collectors.
Determination of vancomycin-resistant enterococci
For the counts of enterococci, we diluted the samples and spread plate them on Slanetz-Bartley agar with and without vancomycin (6 μgmL-1) (Merk, Germany). After incubation during 48 h at 37oC, characteristic enterococci colonies were selected. When the counts were low, the sample was concentrated by filtering through a 0.45 µm membrane (Millipore, U.S). The characteristic colonies were subjected to assays of catalase, bile esculin, and PYR (pyrrolidone arylamidase). Identification was carried out by means of the API 20 Strep system (Biomerieux, France).
Resistance to antibiotics
The study of susceptibility to antibiotics was performed following the Standard for Antimicrobial Susceptibility Testing.8 The minimum inhibitory concentration (MIC) of the VRE was determined by the agar dilution method (serial 2-fold dilutions), for the antibiotics vancomycin (Va) and ampicillin (Ap). The MIC was defined as the least concentration of antibiotic inhibiting visible growth of the bacteria. The positive control used was E. faecium ATCC 19434. Resistance to erythromycin 15 µg (Em), teicoplanin 30 µg (Tp), gentamicin 120 µg (Gm), and tetracycline 30 µg (Tc) (Oxoid, U.K) was determined by the antibiotic disc diffusion method and interpreted according to CLSI criteria.7,10
Statistic analysis
Statistical analyses of the data were performed using the software package IBM SPSS Statistics version 20 (IBM Corp., New York, U.S).
Results
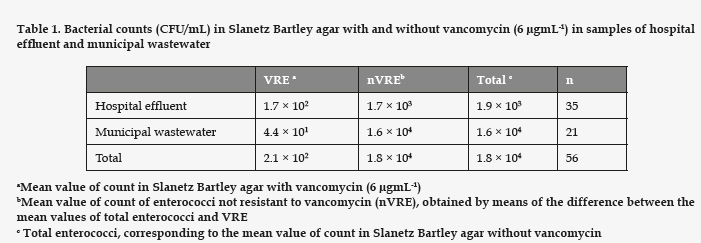
In our study, although enterococci were detected in the samples from both sources, VRE were found in 30% of the samples from the hospital effluent and in 75% of the samples of municipal wastewater. The total counts of enterococci in the urban wastewater were higher than in the hospital effluent (Table 1), giving significant differences (p = 0.036). However, the values of VRE were greater in the hospital effluent, showing an odds ratio of 36.4 (95% CI: 26.0-50.8; p<0.0001).
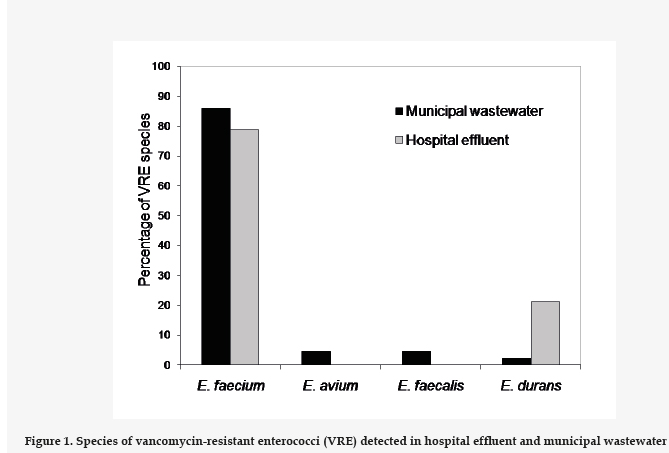
From the samples with VRE, we selected a number of colonies for further studies regarding proportionality with respect to the VRE count. To these end 32 strains from the hospital effluent and 46 strains of municipal wastewater were isolated. Of these, 60 were identified as E. faecium, 2 as E. faecalis, 2 as E. avium, and 6 were E. durans (Fig. 1). Both sources showed a high prevalence of E. faecium.
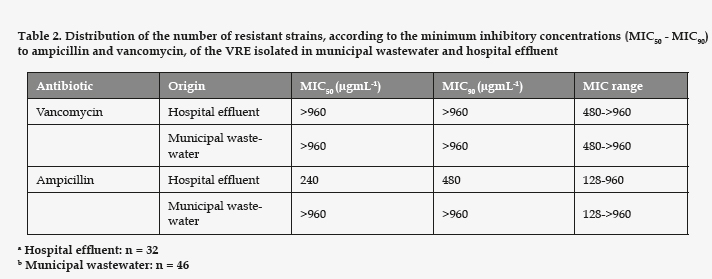
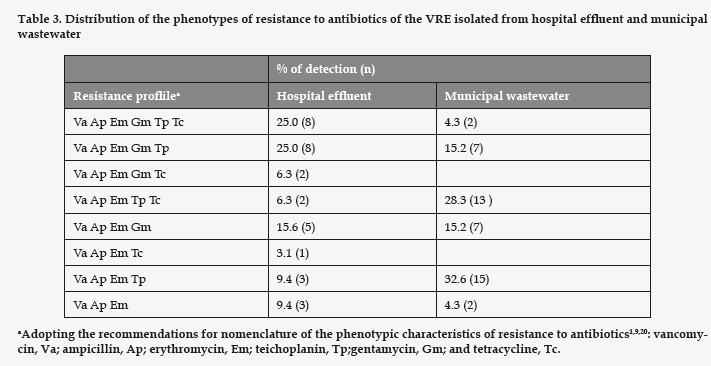
All the strains of enterococci detected in the assays with vancomycin presented high MIC for vancomycin and ampicillin (Table 2). Regarding the rest of the antibiotics tested (Table 3), all the strains studied proved resistant to erythromycin. The VRE strains resistant to gentamicin had a significantly higher prevalence in the hospital effluent (p= 0.008). However, the ones resistant to tetracycline and teicoplanin showed no significant differences between the hospital effluent and the municipal wastewater (p= 0.061 and 0.95, respectively). In the samples of hospital effluent, 25% of VRE were resistant to all six antibiotics assayed. In the samples of municipal wastewater, only 4% were resistant to the six antibiotics, and the most frequent phenotype of resistance was Va Ap Em Tp, identified in 32% of cases.
Discussion
The increase in enterococci with multiple antibiotic resistance stands as a clinical-therapeutic challenge not only in the U.S. and Europe, but in most of the world, including Argentina. Within its genus, Enterococcus faecium is the species most often related with the resistance to multiple antibiotics, vancomycin among them.6 Marín et al.17 described the first clinical isolation of E. faecium resistant to vancomycin in the city of Mendoza (Argentina), attributing its resistance to the gene vanA.
In a previous study, conducted between 1996 and 2010, a total of 1873 isolations of Enterococcus spp. proceeding from hospital infections in the Hospital Universitario José de San Martín (Buenos Aires) were analyzed.22 The prevalence of E. faecium was found to increase from 1.5% in the year 1996 to 4% in 2010, and over 70% of the isolations of E. faecium in hospital infections are resistant to vancomycin, in line with our results (Fig. 1), which would explain the high prevalence of VRE in the hospital effluent of our study. The calculated odds ratio indicates that the presence of VRE is 36.4 times greater in the hospital effluent than in the municipal wastewater. The increased presence of resistant bacteria in hospital effluents and their influence in municipal wastewaters have been described previously, underlining the need to subject hospital effluents to pretreatment.19
Currently, community infections involving VRE have given rise to environmental diffusion, leading to widespread distribution in urban effluents. Most previous studies coincide generally with our results. Goldstein et al.12 found VRE in the influents of four municipal wastewater treatment plants in the U.S., in mean concentrations that ranged between 2.5×103 and 8.6×104 CFU/mL. A study in the city of Porto (Portugal), again involving VRE in a hospital effluent23, arrived at findings in line with ours: the counts of enterococci are similar, and the prevalence of VRE was greater in the hospital effluent than in the municipal wastewater studied. However, our results also show that E. faecium and E. durans are the species of VRE isolate from the hospital effluent, whereas the these authors found E. faecalis to be more frequent.
All research studies to date indicate that the vancomycin resistance in enterococci is most often produced by the gene vanA, characterized by a high resistance to vancomycin and teicoplanin. Beier et al..3 traced 49 of 50 VRE isolates from municipal wastewater to vanA. Our results differ, as only 40.7% of the strains isolated from the hospital effluent, and 32.6% of those proceeding from municipal wastewater were resistant to vancomycin and teicoplanin, implying a lesser prevalence of the vanA phenotype. Yet our results coincide in that 100% of all the VRE strains isolated were resistant to erythromycin, and 34% of those isolated from the municipal wastewater were resistant to tetracycline. These authors found 76% of the strains to be resistant to gentamycin, as opposed to the 34% of our strains of municipal origin.
It is also important to point out that all our strains were ampicillin resistant, possibly due to the presence of β-lactamase. As can be seen in table 2, the MIC was, in all the strains, much higher than 16 µgmL-1 for ampicillin and 32 µgmL-1 for vancomycin, the breakpoints established for resistance.8 The resistance to β-lactam antibiotics is common in VRE strains, as the literature shows. In a South American study involving 20 medical centers, with 218 enterococci (either E. faecalis or E. faecium), 14% of the enterococci were reportedly resistant to vancomycin, the pattern of resistance indicating the gene vanA, and 24.8% were resistant to ampicillin and amoxicillin/clavulanate.14
Conclusion
In conclusion, strains of VRE are found in both hospital effluent and urban wastewater, although the greater concentration would be in hospital effluent. All the VRE isolated in this study were moreover resistant to ampicillin and erythromycin, and the strains of hospital origin presented a greater degree of multi-resistance. Hospital effluent constitutes a source of enterococci having multiple resistance to antibiotics, presumably from the feces of patients, because the rules of biosecurity in medical centers would impede other sources of contamination. The multiresistance patterns found in the VRE strains of municipal wastewater present a different distribution, but their high frequency suggests a selective effect of resistance due to an excessive use of antibiotics in the community. Hospital wastewater increased the health and environment risk of sewage, contributing to the dissemination of bacteria resistant to biocides.
Fundings
The authors declare that they have no funding
Acknowledgments
We would like to acknowledge César Criado Sánchez, Laboratory Technician of the Department of Preventive Medicine and Public Health, Faculty of Pharmacy, University of Granada, Spain, for his invaluable help in making this study possible. We also thank Jean Louise Sanders for translating and editing the manuscript.
Competing interest
The authors declare that they have no conflicts of interest.
References
1. Bachmann BJ, Low KB. Linkage map of Escherichia coli K12, edition 6. Microbiol Rev. 1980; 44 (1):1-56. [ Links ]
2. Baquero F, Martínez JL, Cantón R. Antibiotics and antibiotic resistance in water environments. Curr Opin in Biotechnol. 2008; 19:260-265. [ Links ]
3. Beier RC, Duke SE, Ziprin RL, Harvey RB, Hume ME, Poole TL, Scott HM, Highfield LD, Alali WQ, Andrews K, Anderson RC, Nisbet DJ. Antibiotic and disinfectant susceptibility profiles of vancomycin-resistant Enterococcus faecium (VRE) isolated from community wastewater in Texas. Bull Environ Contam Toxicol. 2008; 80: 188-194. [ Links ]
4. Bonten MJM, Willems R, Weinstein RA. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect Dis. 2001; 1: 314-325. [ Links ]
5. Castillo-Rojas G, Mazari-Hiriart M, Ponce de Leon S, Amieva-Fernández RI, Agis-Juarez RA, Huebner J, López-Vidal Y. Comparison of Enterococcus faecium and Enterococcus faecalis strains isolated from water and clinical samples: antimicrobial susceptibility and genetic relationships. PLoS One.2013; 8(4): e59491. [ Links ]
6. Cetinkaya, Y., Falk, P. and Mayhall, C.G. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000; 13: 686-707. [ Links ]
7. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 17th Informational Supplement, 2007; M100-S17. Wayne, PA, EE.UU. [ Links ]
8. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 22nd Informational Supplement, 2012; M100-S22. Wayne, PA, EE.UU. [ Links ]
9. Demerec M, Adelberg EA, Clark AJ, Hartman PE. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966; 54(1): 61-76. [ Links ]
10. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters, version 5.0. 2015. (On-line) http://www.eucast.org/clinical_breakpoints/. [ Links ]
11. Farrell DJ, Morrissey I, De Rubeis D, Robbins M, Felmingham D.A UK multicenter study of the antimicrobial susceptibility of bacterial pathogens causing urinary tract infection. J Infect. 2003; 46: 94-100. [ Links ]
12. Goldstein RER, Micallef SA, Gibbs SG, George A, Claye E, Sapkota A, Joseph SW, Sapkota AR. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Science of the Total Environment. 2014; 466-467: 404-411. [ Links ]
13. Harwood VJ, Delahoya NC, Ulrich RM, Kramer MF, Whitlock JE, Garey JR, Lim DV. Molecular confirmation of Enterococcus faecalis and E. faecium from clinical, faecal and environmental sources. Letters in Applied Microbiology. 2004; 38:476-482. [ Links ]
14. Jones RN, Guzmán-Blanco M, Gales AC, Gallegos B, Leal Castro AL, Valle Martino MD, Vega S, Zurita J, Cepparulo M. Castanheira M. Susceptibility rates in Latin American nations: report from a regional resistance surveillance program (2011). Braz J Infect Dis. 2013; 17: 672-681. [ Links ]
15. Kummerer K. Henninger A. Promoting resistance by the emission of antibiotics from hospitals and households into effluent. Clin Microbiol Infect. 2003; 9:1203-1214. [ Links ]
16. Leclercq R, Oberlé K, Galopin S, Cattoir V, Budzinski H, Petit F. Changes in enterococcal populations and related antibiotic resistance along a medical center-wastewater treatment plant-river continuum. Appl Environl Microbiol. 2013; 79:2428-2434. [ Links ]
17. Marin ME, Mera JR, Arduino RC, Correa AP, Coque TM, Stamboulian D, Murray BE. First report of vancomycin-resistant Enterococcus faecium isolated in Argentina. Clin Infect Dis, 1998; 26: 235-236. [ Links ]
18. Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut. 2009; 157: 2893-2902. [ Links ]
19. Novo A, Manaia CM. Factors influencing antibiotic resistance burden in municipal waste water treatment plants. Appl Microbiol Biotechnol. 2010; 87: 1157-1166. [ Links ]
20. Novick RP, Clowes RC, Cohen SN, Curtiss III R, Datta N, Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976; 40: 168-189. [ Links ]
21. Paz, M., Muzio, H., Gemini, V., Magdaleno, A., Rossi, S., Korol, S. and Moretton, J. Aguas residuales de un Centro Hospitalario de Buenos Aires, Argentina: Características químicas, biológicas y toxicológicas. Hig Sanid Ambient. 2004; 4: 83-88. [ Links ]
22. Rodríguez CH, García S, Barberis C, Saposnik E, Weyland B, Nastro M, Famiglietti A. Enterococcus spp.: Resistencia antimicrobiana en infecciones intrahospitalarias. Acta Bioquím Clín Latinoam. 2013; 47: 155-160. [ Links ]
23. Varela AR, Ferro G, Vredenburg J, Yanik M, Vieira L, Rizzo L, Lameiras C, Manaia CM. Vancomycin resistant enterococci: From the hospital effluent to the urban wastewater treatment plant. Science of the Total Environment. 2013; 450-451:155-161. [ Links ]
24. Werner G. (2012). Current trends of emergence and spread of vancomycin-resistant enterococci. En: Pana M, editors. Antibiotic resistant bacteria - A continuous challenge in the new millennium. Croatia, In Tech, 2012, p. 303-354. [ Links ]
25. Willems RJL, Top J, Van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJM. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis. 2005; 11:821-828. [ Links ]
![]() Correspondence:
Correspondence:
Elena Moreno
elmorol@ugr.es
Received: 12-09-2016
Accepted: 29-09-2016