My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Archivos de Zootecnia
On-line version ISSN 1885-4494Print version ISSN 0004-0592
Arch. zootec. vol.63 n.241 Córdoba Mar. 2014
https://dx.doi.org/10.4321/S0004-05922014000100024
NOTA BREVE
Microsatellite characterization of the Momil, Cordoba (Colombia) domestic pig
Caracterización mediante microsatélites del cerdo doméstico en Momil, Córdoba
Pardo, E.1*; Cavadía, T.1A and Meléndez, I.2
1Area of Genetics. Biology Department. University of Córdoba. Montería. Colombia. *epardop@correounicordoba.edu.co; Atcavadia@correo.unicordoba.edu.co
2Area of Genetics. Biology Department. University of Pamplona. Pamplona. Colombia. imgelvez@hotmail.com
SUMMARY
The genetic status of the domestic pig (Sus scrofa domestica) from Momil, Colombia, was studied on 45 samples using 20 microsatellites recommended by the FAO/ISAG for swine biodiversity studies. All microsatellites were polymorphic having from 2 (SW1067) to 13 (SW957) alleles. An average of 5.2 and a total of 104 alleles were detected. The average expected heterozygosity was 0.5376 and the observed was 0.5216. PIC values ranged between 0.1689 (SW1067) and 0.7352 (SW957), respectively. The population seems be genetically stable.
Key words: Genetic variation. Hardy-Weinberg equilibrium. Probability of exclusion. Sus scrofa domestica.
RESUMEN
Se estudió la situación genética del cerdo doméstico (Sus scrofa domestica) de Momil, Córdoba, Colombia sobre 45 muestras empleando 20 microsatélites de los recomendados por la FAO/ ISAG para estudios de biodiversidad porcina. Todos los microsatélites resultaron polimórficos y se han detectado, entre 2 (SW1067) y 13 (SW957) alelos, con un número medio de 5,2 y un total de 104. la heterocigosidad media esperada ha sido 0,5376 y la observada 0,5216. El PIC osciló entre 0,1689 (SW1067) y 0,7352 (SW957). La población estudiada es genéticamente estable.
Palabras clave: Variación genética. Equilibrio Hardy-Weinberg. Probabilidad de exclusión. Sus scrofa domestica.
Introduction
In Córdoba (Colombia) the pig (mixtures of different races, ICA, 2013) production, is usually characterized by malnutrition, and bad health management. Animals usually remain on pasture, with free access to the farmers' home. Their feeding in the growth phase (from 20 to 24 months age), is especially based on the intake of food scraps and leftover harvest (banana, yucca, yam, etc). For these reasons they grow weak, and vulnerable to several illnesses, resulting in a low weight. Taking into account that genetic information from samples of domestic pigs (Sus scrofa domestica) in the Caribbean countries, is low, it is necessary to generate studies in order to identify their genetic status. Although, the study of genetic characterization of domestic pigs in Momil, Cordoba is a major challenge from a scientific and strategic point of view, it is an outstanding opportunity to learn, and to have a better understanding of the reality of this species. This type of research will confirm the obligation of research to deepen the knowledge of these genetic resources, and their cultural and economic importance for the region. In this study, a sample of 45 domestic pigs in Momil (Córdoba) was genetically characterized using microsatellite markers in order to identify their genetic status.
Materials and methods
Hair samples were collected in Momil, Cordoba (9o 14 '16 " N and 75o 36' 30" W), Colombia, from 45 specimens. As animals from family farms, there are no genealogy data available. Twenty microsatellites recommended by the FAO/ISAG for genetic diversity studies in pigs were used. For the analysis of fragments and allelic characterization, Genescan Analysis® 3.1.2 and Genotyper® 2.5.2 programs were used respectively. By using the Genetix v 4.02 program (Belkhir et al., 2003), allele frequencies, heterozygosities and FIS value were calculated. The presence of the Hardy-Weinberg equilibrium (HW) was measured with the Genepop v. 3.1c program (Guo and Thompson, 1992). The polymorphic information content (PIC) was calculated with the Ben Hui Liu (1987) algorithm. The allelic richness and FIS values were obtained with the Fstat program.
Results and discussion
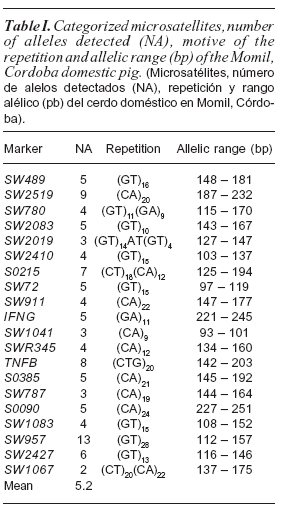
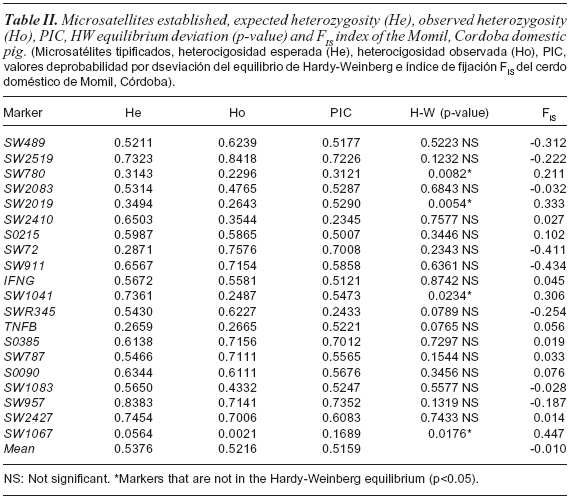
The results show a high degree of polymorphism for all microsatellites, detecting a total of 104 alleles, ranging from 2 (SW1067) and 13 (SW957) alleles per locus (table I) and a number of alleles found in the sample of 5.2 (table I). Other studies of genetic diversity in pigs reported higher and lower values between 2.3 ((Nidup et al., 2011) and 13.31 alleles (Chang et al., 2009). The obtained polymorphic information content (table II) varied between 0,1689 (SW1067) and 0.7352 (SW957). These values correspond to the markers that showed the lowest and the highest number of alleles respectively. Only 16 markers can be considered very informative (PIC>0.7), making them very revealing in detecting the genetic variation in this sample. One marker was moderately informative (PIC>0.25) and 3 not very informative (PIC <0.25). Compared with previously published data, we found that the average value of PIC was lower than that reported by Vicente et al. (2008), and similar to those reported by Castro et al. (2007). Sixteen out of the 20 microsatellites analyzed were found in the Hardy-Weinberg equilibrium, therefore, we can say that the sample is genetically stable (table II). This could show that intra-population mating occurred randomly (with respect to the markers considered) or, if new animals have recently joined this sample, they have come from samples with the same gene pool of the analyzed sample in this study Four loci showed a significant deviation from the Hardy-Weinberg equilibrium (SW2019, SW1041, SW180 and SW1067), revealing an excess of homozygotes. Excess of homozygotes in a sample could be the result of inbreeding events within the sample (Allendorf and Luikart, 2007). However, inbreeding equally affects the entire genome, then, as it is expected, all used markers would show an excess of homozygotes, which did not occur in this case. On the other hand, there might be a genetic structure by subdivision (Wahlund effect). If this is true, it would mean that there are marked differences between close domestic pig samples to the markers SW2019, SW1041, SW180 and SW1067, but not for the other markers. If the differences for these markers were not removed it is because the gene flow between near samples is limited (aspect not shown for the other markers). These differences may also indicate that the markers SW2019, SW1041, SW180 and SW1067, are linked to genes under natural selection, which acts differentially at the micro or macro spatial. Another possibility is the presence of null alleles at these loci, frequent event in much of microsatellite studies (Dakin and Avise, 2004). Null alleles can be caused by high mutation rate that exhibit these types of markers, causing an alteration of the sequence between slightly distant populations, or due to technical problems during PCR. Levels of observed heterozygosity and expected heterozygosity found in the sample studied (table II) show low genetic variability, perhaps due to consanguineous mating, something often carried out by animal owners. The percentage of heterozygotic individuals was above 50 %, reaching values of 52.16 % for the mean observed heterozygosity and 53.76 % for the expected average heterozygosity. These values are higher than those reported by Oslinger et al. (2006) in Colombia and above those reported in India, Thailand and Spain (Nidup et al., 2011). The mean number of alleles per locus and allelic richness indicate that this sample exhibits a certain degree of variability. The FIS value found demonstrates a homogeneous sample in its genetic characteristics.
Conclusions
The chosen microsatellites contribute to the genetic characterization of this sample. High polymorphism, low genetic variability, and high PIC values were observed, resulting in very informative and useful markers to measure the genetic diversity. The usefulness of this study will increase when analyzing other populations using phylogenetic mating, especially to contrast and clarify genetic relationships with the Momil pigs.
References
1. Allendorf, F.W. and Luikart, G. 2007. Conservation and the genetics of populations. Blackwell. Maiden, Massachusetts. USA. [ Links ]
2. Belkhir, K.; Borsa P.; Chikhi, L.; Raufaste, N. et Bonhomme, F. 2003. Genetix: 4.02 Logiciel sous Windows™ pour la genetique des populations. Laboratoire Génome, Populations, Interactions. Adaptations. Université de Montpellier. Montpellier. France. [ Links ]
3. Ben Hui Liu. 1987. Statistical Genomics. CRC Press. Raleigh, North Carolina. USA. [ Links ]
4. Castro, G.; Santandreu, A.; Ronca, F.; y Lozano, A. 2007. La cría de cerdos en asentamientos urbanos y periurbanos de Montevideo (Uruguay). "Porcicultura urbana y periurbana en ciudades de América Latina y el Caribe". Serie Cuadernos de Agricultura Urbana. 1a ed. Lima. Perú. pp. 34-39. [ Links ]
5. Chang, W.H.; Chu, P.H.; Jiang, Y.N.; Li, S.H.; Wang, Y.; Chen, C.H.; Chen, K.J.; Lin, C.Y. and Ju, Y.T. 2009. Genetic variation and phylogenetics of Lanyu and exotic pig breeds in Taiwan analyzed by nineteen microsatellite markers. J Anim Sci, 87: 1-8. [ Links ]
6. Dakin, E.E. and Avise, J.C. 2004. Microsatellite null alleles in parentage analysis. Heredity, 93: 504-509. [ Links ]
7. Guo, S. and Thompson, E. 1992. Performing the exact test of Hardy-Weinberg proportions for multiple allele. Biometrics, 48: 361-372. [ Links ]
8. ICA. 2013. Censo pecuario nacional Colombia. http://www.ica.gov.co/getdoc/8232c0e5-be97-42bd-b07b-9cdbfb07fcac/Censos-2012.aspx (18/02/2013). [ Links ]
9. Nidup, k. and Moran, C. 2011. Genetic diversity of domestic pigs as revealed by microsatellites: A mini review. Genom Quant Genet, 2: 5-18. [ Links ]
10. Oslinger, A; Muñoz, E.J.; Álvarez, L.A.; Ariza, F.; Moreno, F. y Posso, A. 2006. Caracterización de cerdos criollos colombianos mediante la técnica molecular RAMs. Acta Agron, 55: 45-50. [ Links ]
11. Vicente, A.A.; Carolino, M.I. and Sousa, M.C. 2008. Genetic diversity in native and specialized breeds of pigs in Portugal assessed by microsatellites. J Anim Sci, 86: 2496-2507. [ Links ]
Recibido: 14-5-13
Aceptado: 12-6-13