Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Archivos de Zootecnia
versión On-line ISSN 1885-4494versión impresa ISSN 0004-0592
Arch. zootec. vol.58 no.224 Córdoba dic. 2009
Mitochondrial DNA D-loop analysis of South Western Nigerian chicken
Analisis de D-Loop ADN mitocondrial de pollos de SW Nigeria
Adebambo, A.O.1 and the Chicken Diversity Consortium*
1Department of Animal Breeding and Genetics. University of Agriculture. Abeokuta, P.M.B. 2240. Abeokuta. Naigeria. tumininuadebambo@yahoo.com
*Chicken Diversity Consortium (Africa): G. Bjørnstada, W. Bulimob, H. Jianlina, G. Kiersteina, L. Mazhanid, B. Podisid, J. Hirboa, K. Agyemangc, C. Wollnye, T. Gondwel, V. Zeuhf, D. Tadelleg, G. Abebeg, P. Abdoulayeh, S. Pacoi, L. Serunjogij, M. Abrerrahmanf, R. Sowh, S. Weigendm, R. Sanfoi, F. Gayec, E. Ssewanyanaj, M. D. Coulibalyk, B. Temek, VSF (Sudan) and O. Hanottea.
aInternational Livestock Research Institute (ILRI). P.O. Box 30709. Nairobi 00100. Kenya.
bDepartment of Biochemistry. University of Nairobi. P.O. Box 30197. Nairobi 00100. Kenya.
cInternational Trypanotolerance Centre (ITC). The Gambia.
dAgricultural Research. Private Bag 0033. Gaborone. Botswana.
eInstitute of Animal Breeding. Georg-August-Universität Göttingen. Germany.
fProject de Développement Rural de la Préfecture du Lac (PDRPL). B.P. 782. N'Djamena. Tchad.
gInternational Livestock Research Institute (ILRI). P.O. Box 5689. Addis Ababa. Ethiopia.
hInstitut Sénégalais de Recherches Agricoles. Dakar. Sénégal.
iInstitut de l'Environnement et de Recherches Agricoles (INERA). Ouagadougou. Burkina Faso.
jSerere Agricultural and Animal Production Research Institute. Privatebag. Soroti. Uganda.
kInstitut d'Economie Rurale. P.O Box 258. Rue Mohamed V. Bamako. Mali.
lDepartment of Animal Science. University of Malawi. Bunda College of Agriculture. Malawi.
mInstitute for Animal Breeding. Federal Agricultural Research Centre. Mariensee. Germany.
SUMMARY
Mitochondrial DNA (mtDNA) D-loop segment was sequenced for a total of 98 individuals of domestic chicken from South Western Nigeria. Domestic chicken populations were: Anak titan (Israeli breed,n= 1), Frizzle (n= 16), Opipi (n= 5), FrizzleXOpipi (n= 5), Fulani (n= 4), Giriraja (Indian breed,n= 3), Normal (n= 55), Naked neck (n= 8), Yaffa (n= 1). The sequences of the first 397 nucleotides were used for the analysis. Seventeen haplotypes were identified in the samples, 15 for Nigerian indigenous chicken population, 1 for Giriraja and 1 for Anak titan from 23 polymorphic sites. Phylogenetic analysis shows that Nigerian indigenous and Anak titan chicken were all grouped under clade IV, while the Indian Giriraja was under clade IIIc. Clade IV had 16 haplotypes, while clade IIIc had one haplotype. AMOVA analysis indicates that 97.32% of the total sequence variation between haplotypes was present within population and 2.68% between populations. Our results suggest single multiple maternal origins for the South Western Nigerian domestic chicken.
Key words: Local poultry breeds. Genetic diversity. Haplotype. Phylogenetic tree.
RESUMEN
Un segmento D-loop de AND mitocondrial mtADN) fue secuenciado para un total de 98 pollos domésticos de SW Nigeria. Las poblaciones domésticas de pollos fueron: Anak titan (raza israelí, n= 1), Frizzle (n= 16), Opipi (n= 5), FrizzlexOpipi (n= 5), Fulani (n= 4), Giriraja (raza india, n= 3), Normal (n= 55), Cuello Desnudo (n= 8), Yaffa (n= 1). Las secuencias de los primeros 397 nucleotidos fueron usadas para el análisis. Diecisiete haplotipos de 23 sitios polimórficos, fueron identificados en las muestras: 15 para las poblaciones indígenas nigerianas de pollos, 1 para Giriraja y 1 para Anak Titan. El análisis filogenético, muestra que los pollos indígenas nigerianos pueden agruparse todos dentro del clade IV, mientras que el Giriraja indio, se encuadró en el clade IIIc. El clade IV tiene 16 haplotipos mientras que el clade IIIc tiene sólo un haplotipo. El análisis AMOVA indica que el 97,32% de la variación total de la secuencia entre haplotipos estuvo presente dentro de la población y el 2,68% entre poblaciones. Los resultados sugieren un solo origen maternal múltiple para los pollos domésticos de SW Nigeria.
Palabras clave: Razas aviares locales. Diversidad genética. Haplotipo. Árbol filogenético.
Introduction
It is widely established that all populations of domesticated chicken descend from a single ancestor, the red jungle fowl (Gallus gallus gallus), which originated in Southeast Asia (Akishinonomiya et al., 1994, 1996). Chicken is the most widely distributed of all livestock and poultry species in African countries. It plays a very significant role as a source of income and high quality protein to the rural households.
Knowledge on the distribution of chicken genetic diversity in Africa would be useful in optimizing both conservation and utilization strategies for indigenous chicken genetic resources. The diversity of Nigerian local chickens reported is based mostly on phenotypes including plumage color, feathering pattern, adult body weight, egg weight, reproduction performance and immune responses to various diseases (Omeje and Nwosu, 1983, Nwosu et al., 1985, Ikeobi et al., 1996, Adebambo et al., 1999, Adebambo, 2002) which have limited utility in the study of genetic variation. Mitochondrial DNA (mtDNA) sequences have successfully been used to determine genetic diversity in Asian chicken (Niu et al., 2002; Liu et al., 2004) and African chicken (Mobegi and Chicken Diversity Consortium, 2005).
Chicken mtDNA has 16,775 base pairs (Desjardins and Morais, 1990). MtDNA is highly polymorphic compared to nuclear DNA, evolutionary rate being 5 to 10 times faster than the nuclear genome (Brown et al., 1982). Different regions of the mtDNA evolve at different rates (Saccone et al., 1991), making it a marker of choice for studying genetic diversity within as well as between species. The displacement (D)-loop region is non-coding and evolves much faster than other regions of the mtDNA genome. This makes it particularly useful for phylogeographic analysis (Avise, 1994). MtDNA is maternally inherited in most species and does not undergo recombination (Hayashi et al., 1985). These features mean that each molecule as a whole usually has a single genealogical history through maternal lineage.
In the present study, the sequences of the D-loop hypervariable 1 (HV1) segment of the mtDNA were used to study the genetic diversity and relationship of South Western Nigerian domestic chicken.
Materials and methods
Chicken blood samples from a total of 98 individuals belonging to 3 major phenotypes were collected from South-western Nigeria using FTA® classic cards (Whatman BioScience, Maidstone, UK). Geographic location and number of the samples collected were as follow: Lagos (n= 7, Epe), Ogun (n= 20, Imeko, Ipokia, Owode), Oyo (n= 15, Ago-are, Oke-iho), Osun (n= 7, Ifewara), Ondo (n= 16, Ikare, Ore), Kwara (n= 24, Jebba, Kosubosu, Offa) Edo (n= 4, Uromi) and University of Agriculture, Abeokuta (n= 5, exotic chicken stocks). Genomic DNA was extracted from air-dried blood spotted on filter paper (FTA® classic cards) following the recommended manufacturer's protocol. The primers used to amplify the hypervariable 1 (HV1) segment were L16750 (5'AGGACTACGGCTTGAAAAGC-3') as forward primer and H547 (5'ATGTGCCTGA CCGAGGAACCAG-3') as reverse primer. This primer pair amplifies a 550bp fragment between sites 16750 (GenBank accession number NC_001323, Desjardins and Morais, 1990) and 547 (GenBank accession number AB098668, Komiyama et al., 2003). PCR reactions were performed in a 30 μl reaction volume containing 2.5 mM of each dNTPs, 14 pmol of each primer, 1.5 mM MgCl2, 1 x PCR buffer comprising 10 mM Tris-HCl (pH 8.3) and 50 mM KCl, and 1.25 U Taq DNA polymerase (Promega, Madison, USA). PCR amplifications were carried out on a GeneAmp® PCR system 9700 (Applied Biosystems, USA) thermal cycler. The reaction profile was: initial denaturation at 94oC for 2 min, followed by 35 cycles at 94oC for 30 s, 58oC for 30 s and 72oC for 1 min. The last cycle was followed by a final extension step at 72oC for 10 min. PCR products were electrophoresed on a 1.5% (w/v) agarose gel stained with ethidium bromide in a 1 x TBE buffer at 100 volts for 1 hour. PCR products were purified using the QIAquick PCR purification kit (QIAGEN, GmbH, Germany) according to the manufacturer's protocol. Direct sequencing of HV1 segment of the D-loop region was performed using two internal primers CR-for (5'TCTATA TTCCACATTTCTC-3') and CR-rev (5'-GCGAGCATAACCAAATGG-3'). Sequencing was done using the BigDye® Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems, USA) and the purified sequencing products were electrophoresed on an ABI 3730 XL automated capillary DNA sequencer (Applied Biosystems, USA).
MtDNA sequences for the first 397 nucleotides of D-loop were aligned using the program ClustalX 1.83 (Thompson et al., 1997; available at ftp://ftp-igbmc.ustrasbg.fr/pub/ClustalX). Polymorphic sites were identified with the program DnaNASP (Rozas et al., 2003; available at http://www.ub.es/dnasp). Phylogenetic analyses were conducted using the program MEGA version 3.0 (Kumar et al., 2004; available at http://www.megasoftware.net/). Median-joining network analysis of the haplotypes based on the variable characters of the complete alignment was conducted using computer program NETWORK 4.1.0.8 (Bandelt et al., 1999; available at http://www.fluxus-engineering.com). Maternal genetic differentiation was quantified using hierarchical analysis of molecular variance (AMOVA) (Excoffier et al., 1992; http://anthro.unige.ch/arlequin).
Results and discussion
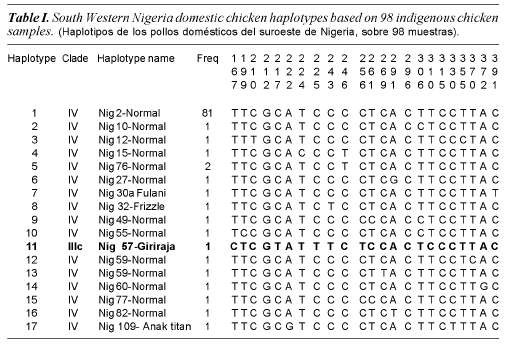
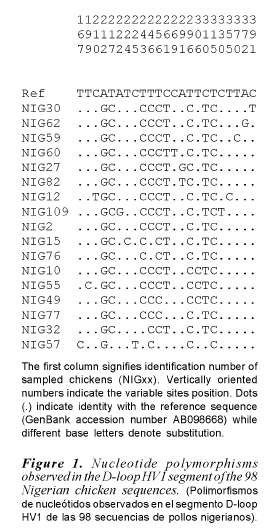
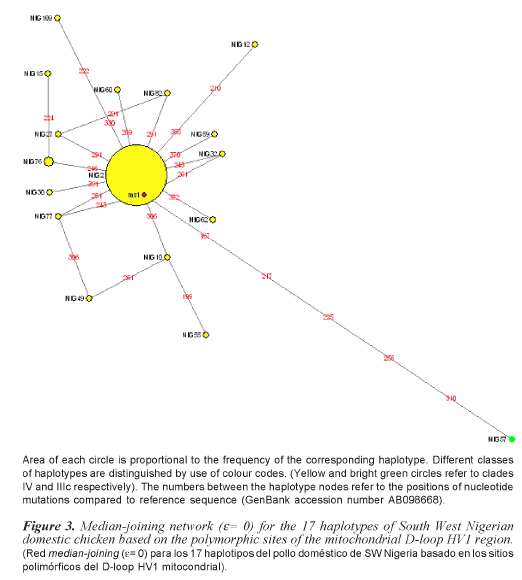
Ninety eight chicken samples collected from South Western Nigeria resulted in 17 haplotypes identified from 23 polymorphic sites. Haplotypes identified in the 98 South Western Nigeria chicken, their clades (haplogroups) and frequencies are shown in table I. Nucleotide polymorphisms observed in the D-loop HV1 segment of the Nigerian chicken sequences are shown in figure 1.
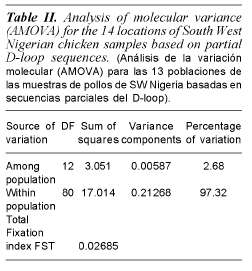
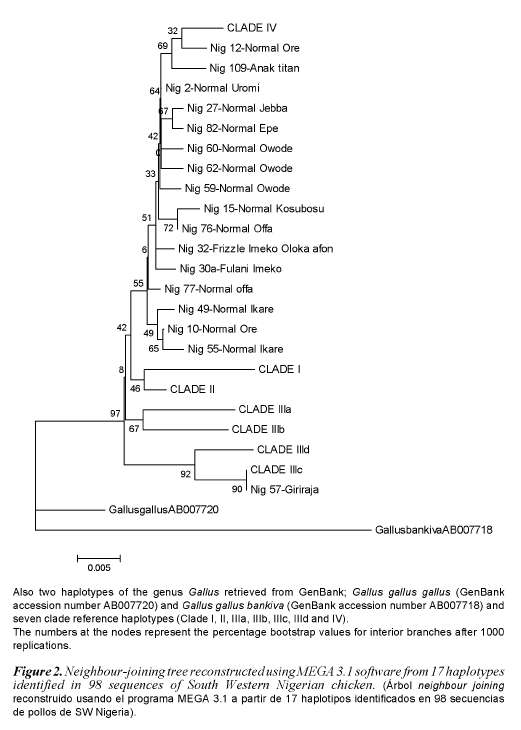
Phylogenetic analysis indicates that south western Nigerian domestic chicken can be grouped into one clade (Clade IV) of the seven clades (Clade I, II, IIIa, IIIb, IIIc, IIId and IV) that were previously identified in Asian domestic chicken (Yi-Ping et al., 2006). A phylogenetic tree with 17 different haplotypes found in Nigerian domestic chicken is shown in figure 2. Analysis of molecular variance analysis (table II) indicates that 97.32% of the total sequence variation between haplotypes was present within population and 2.68% between populations. Median joining networks were drawn for the haplotypes identified in 98 sequences of South Western Nigerian domestic chicken. The network illustrates the relationship between 17 haplotypes (figure 3). There are sixteen haplotypes in clade IV and one in clade IIIc.
Our results indicate that mtDNA (HV1) D-loop region is variable in South Western Nigerian domestic chicken exhibiting 17 haplotypes which indicate multiple maternal origins for the South Western Nigerian domestic chicken.
References
Adebambo, A.O. 2002. Evaluation of the genetic variation among growth traits of Indian and Nigerian chicken genotypes. M. Agric. Dissertation. Department of Animal Breeding and Genetics. University of Agriculture. Abeokuta. 106 pp. [ Links ]
Adebambo, O.A., C.O.N. Ikeobi, M.O. Ozoje and J.A. Adenowo. 1999. Color variations and performance characteristics of the indigenous chicken of South Western Nigeria. Niger. J. Anim. Prod., 26: 15-22. [ Links ]
Akishinonomiya, F., T. Miyake, S. Sumi, M. Takada, S. Ohno and N. Kondo. 1994. One subspecies of the Red Jungle Fowl (Gallus gallus gallus) suffices as the matriarchic ancestor of all domestic breeds. Proc. Natl. Acad. Sci. USA., 91: 12505-12509. [ Links ]
Akishinonomiya, F., T. Miyake, M. Takada, R. Shingu, T. Endo, T. Gojobori, N. Kondo and S. Ohno. 1996. Monophyletic origin and unique dispersal patterns of domestic fowls. Proc. Natl. Acad. Sci. USA., 93: 6792-6795. [ Links ]
Avise, J.C. 1994. Molecular markers: natural history and evolution. Chapman and Hall. New York. [ Links ]
Bandelt, H.J., P. Forster and A. Rohl. 1999. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol., 16: 37-38. [ Links ]
Brown, W.M., E.M. Prager, A. Wang and A.C. Wilson. 1982. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J. Mol. Evol., 18: 225-239. [ Links ]
Desjardins, P. and R. Morais. 1990. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J. Mol. Biol., 212: 599-634. [ Links ]
Excoffier, L., P. Smouse and J. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 131: 479-491. [ Links ]
Hayashi, Y., T. Nishida, T. Fujioka, I. Tsugiyama and K. Mochizuki. 1985. Osteometrical studies on the phylogenetic relationships of Japanese native fowls. Jap. J. Vet. Sci., 47: 25-37. [ Links ]
Ikeobi, C.O.N., M.O. Ebozoje, O.A. Adebambo, J.A. Adenowo and O.A. Osinowo. 1996. Genetic differences in the performance of the local chicken in South Western Nigeria. Niger. J. Genet., 11: 55-60. [ Links ]
Komiyama, T., K. Ikeo and T. Gojobori. 2003. Where is the origin of the Japanese gamecocks? Gene, 317: 195-202. [ Links ]
Kumar, S., K. Tamura and M. Nei. 2004. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics, 5: 150-163. [ Links ]
Liu, Z.G., C.Z. Lei, J. Luo, C. Ding, G.H. Chen, H. Chang, K.H. Wang, X.X. Liu, X.Y. Zhang, X.J. Xiao and S.L. Wu. 2004. Genetic variability of mtDNA sequences in Chinese native chicken breeds. Asian-Aust. J. Anim. Sci., 17: 903-909. [ Links ]
Mobegi, A.V. and Chicken Diversity Consortium. 2005. Mitochondrial DNA D-loop sequences reveal the genetic diversity of African chicken. Proceedings of the 4th All Africa Conference on Animal Agriculture. September 20-24. [ Links ]
Niu, D., Y. Fu, J. Luo, H. Ruan, X.P. Yu, G. Chen and Y.P. Zhang. 2002. The origin and genetic diversity of Chinese native chicken breeds. Biochem. Genet., 40: 163-174. [ Links ]
Nwosu, C.C., F.A. Gowen, F.C. Obioha, I.A. Akpan and G.I. Onuora. 1985. Biometrical study of the conformation of the native chicken. Niger. J. Anim. Prod., 12: 141-146. [ Links ]
Omeje, S.S.I. and C.C. Nwosu. 1983. Egg production patterns in local chickens and their crosses in the short term. Niger. J. Anim. Prod., 10: 91-96. [ Links ]
Rozas, J., J.C. Sanchez-Del Barrio, X. Messeguer and R. Rozas. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19: 2496-2497. [ Links ]
Saccone, C., G. Pesole and E. Sbisa. 1991. The main regulatory region of mammalian mito-chondrial DNA: structure-function model and evolutionary pattern. J. Mol. Evol., 33: 83-91. [ Links ]
Thompson, J.D., T.J. Gibson, F. Plewniak, F. Jeanmougin and D.G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25: 4876-4882. [ Links ]
Yi-Ping, L., W. Gui-Sheng, Y. Yong-Gang, M. Yong-Wang, G. Luikart, M. Baig, A. Beja-Pereira, D. Zhao-Li, M. Gounder Palanichamy and Z. Ya-Ping. 2006. Multiple maternal origins of chickens: out of the Asian jungle. Mol. Phylogen. Evol., 38: 12-19. [ Links ]
Recibido: 10-10-07.
Aceptado: 21-2-08.