INTRODUCTION
The global obesity epidemic continues to grow and now affects more than two billion people, or 30 % of the world’s population (1). Obesity is ranked as the fifth leading cause of death worldwide and is associated with the development of many diseases (2). Body mass index (BMI) is commonly used to define overweight and obesity (3). It has been shown that obesity is associated with a higher risk of breast cancer, while relatively little research has been done on the relationship between obesity and inflammatory breast disease (4).
Inflammatory breast diseases are common benign breast diseases and are classified as lactational mastitis and non-lactational mastitis. Non-lactating mastitis is less common and includes idiopathic granulomatous mastitis, periductal mastitis, and tuberculous mastitis (5). Many previous observational studies have shown a strong association between obesity and mastitis (6,7). From the perspective of evidence-based medicine, the causal relationship between obesity and inflammatory breast disease is unclear. There are few clinical studies on obesity and mastitis and only a few observational studies, which are not convincing enough due to their shortcomings and many confounding factors.
Mendelian randomization (MR) studies have gotten a lot of attention since the recent addition of genome-wide association studies (GWAS) databases. Mendelian randomization (MR), conceptually similar to randomized controlled trials (RCTs), is based on the principle of random assignment of genetic variants at meiosis, which reduces the impact of confounding encountered in observational epidemiology (8-10). MR studies have been widely employed to determine the causal relationship between an exposure and an outcome.
In this study, we investigated the genetic causal relationship between BMI and inflammatory breast disease using MR analysis.
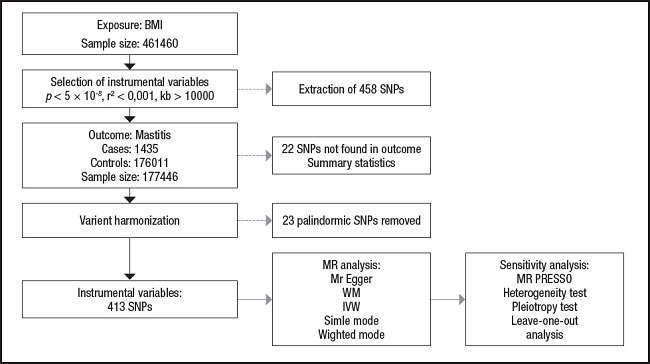
The overview of the research design is shown in figure 1.
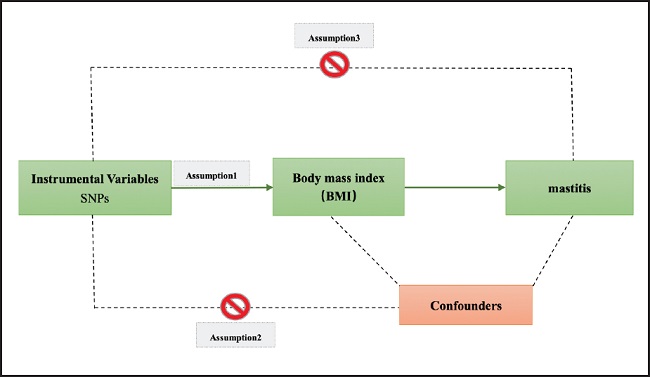
Figure 1. The study assumptions of the two-sample Mendelian randomization analysis between body mass index (BMI) and mastitis. The assumptions included: a) single nucleotide polymorphisms (SNPs) selected as instrumental variables (IVs) should be closely related with exposure; b) selected SNPs must be independent of confounders; and c) IVs are associated to outcome only via exposure rather than through a direct association.
MATERIALS AND METHODS
DATA SOURCE
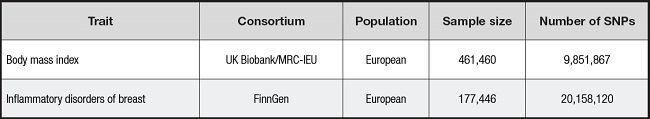
Data on BMI were obtained from the GWAS European Population Summary database supported by the UK Biobank (https://gwas.mrcieu.ac.uk/datasets/ukb-b-19953/). This database contains 461,460 European participants. Data on outcome variables for inflammatory breast disease were obtained from the FinnGen Biobank (https://r8.finngen.fi), which contains 1,435 cases of inflammatory breast disease and 176,011 controls of European ancestry (11).
INSTRUMENT SELECTION CRITERIA
We extracted genetic instrumental variables (IVs) using the following criteria: a) p value (genome-wide significance threshold < 5e-8) (12); and b) in order to avoid the potential bias caused by strong linkage disequilibrium (LD) (13), a linkage disequilibrium correlation coefficient r2 (r2 < 0.001), and a number of bases between two SNPs (kb > 10,000) were set. Next, we combined the exposed and outcome data and removed the palindromic SNPs, leaving the remaining SNPs as the final instrumental variables used for analysis. We then used the F-statistic (F = beta2/se2) to assess the statistical power of the remaining SNPs and calculated the F-values of all SNPs and removed those with low statistical power (F-statistic < 10) (14). If the F value for the instrument exposure correlation was significantly greater than 10, the likelihood of weak instrumental variable bias was low (15).
STATISTICAL APPROACHES AND SENSITIVITY ANALYSES
Data analysis in this study was performed using R (version 4.2.2) through TwoSampleMR (0.5.6) package and the MR-Pleiotropy Residual Sum and Outlier method (MR-PRESSO) (1.0) (16). First, to assess the level of significance of the causal relationship between BMI and mastitis, we used a two-sample MR analysis. In order to assess whether BMI has a causal effect on mastitis, we mostly used the inverse-variance weighted (IVW) method and the MR-Egger method, along with weighted median, simple mode, and weighted mode approaches (17-19). The IVW method is a crucial method for realizing unbiased estimates since it guarantees the necessary statistical power to establish a link between an exposure factor and an outcome. MR-Egger regression is similar to IVW, except that it includes an intercept term for the average pleiotropic effect. The intercept in the MR-Egger regression can be used to test the IV hypothesis (20). Evaluation of potential heterogeneity and horizontal pleiotropy requires sensitivity analysis. Cochran’s Q test was performed to assess the heterogeneity of effect sizes for selected genetic IVs. The MR-PRESSO analysis was also applied to exclude outliers and moderate horizontal pleiotropy (21). The intercept derived from MR-Egger regression was employed to evaluate vertical pleiotropy (22). We also performed a “leave-one-out” sensitivity test as part of our sensitivity analysis to check for significant variations in our models both before and after eliminating each IV (17,23). The flow chart regarding the Mendelian randomization analysis of this study is shown in figure 2.
RESULTS
GENETICS VARIANTS SELECTION
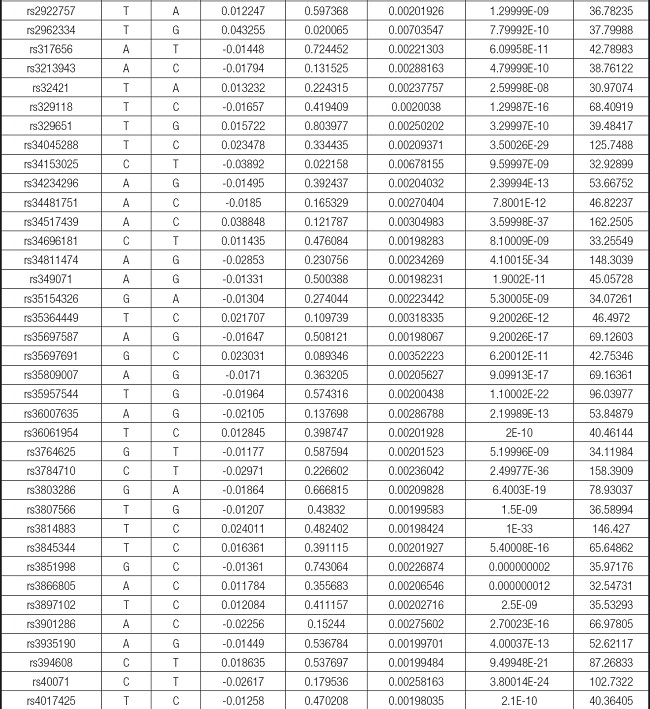
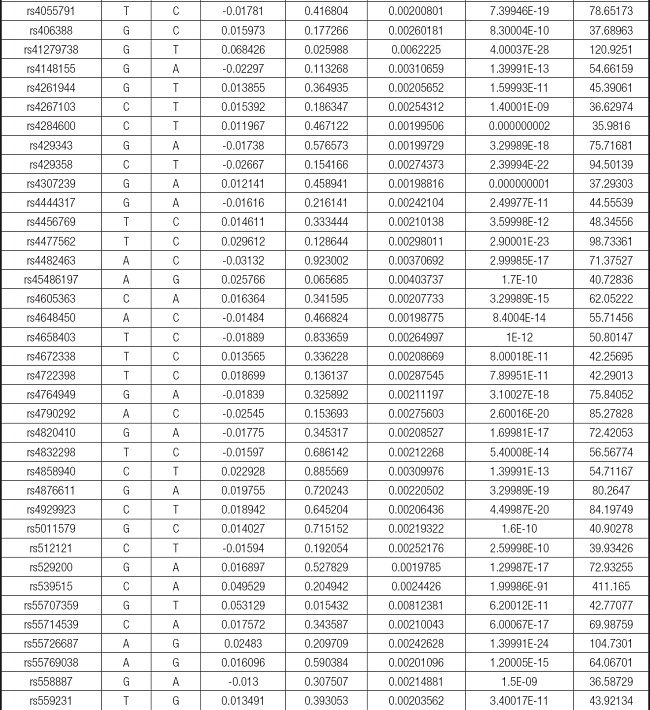
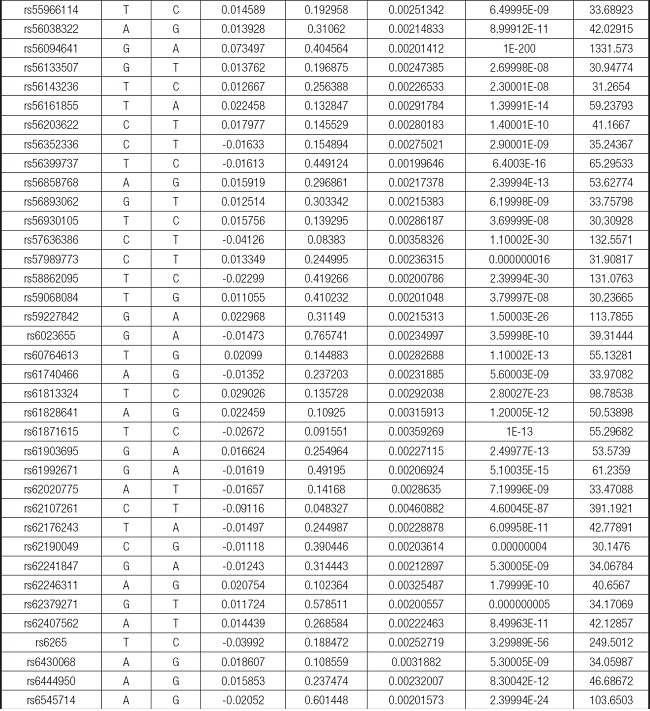
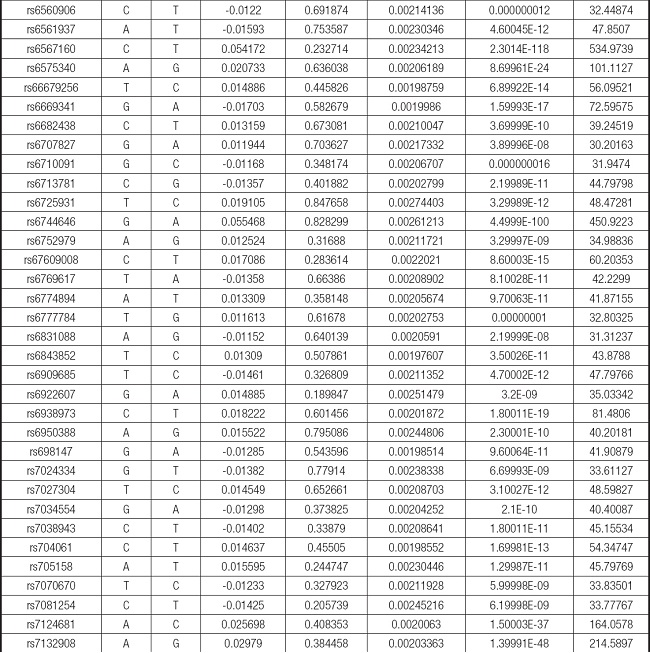
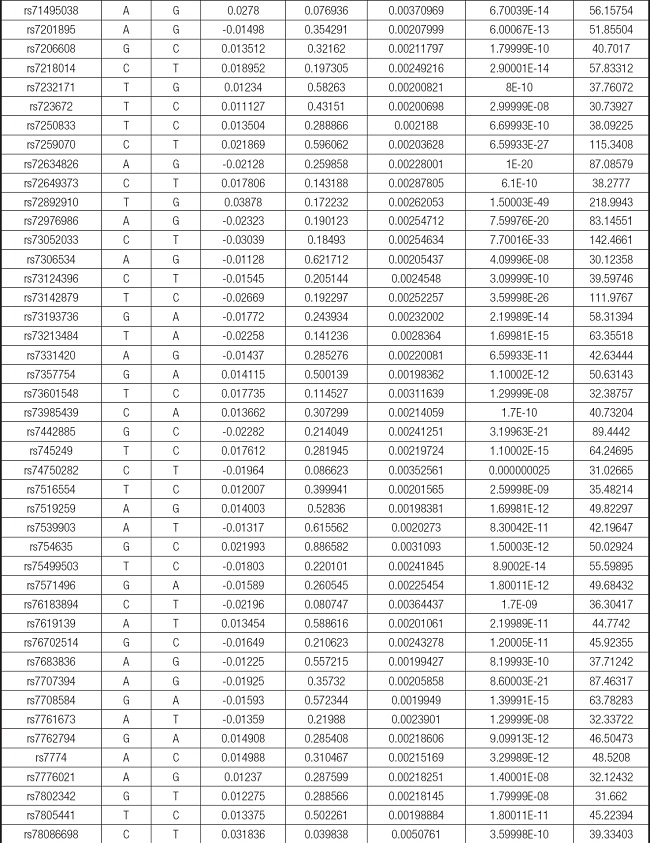
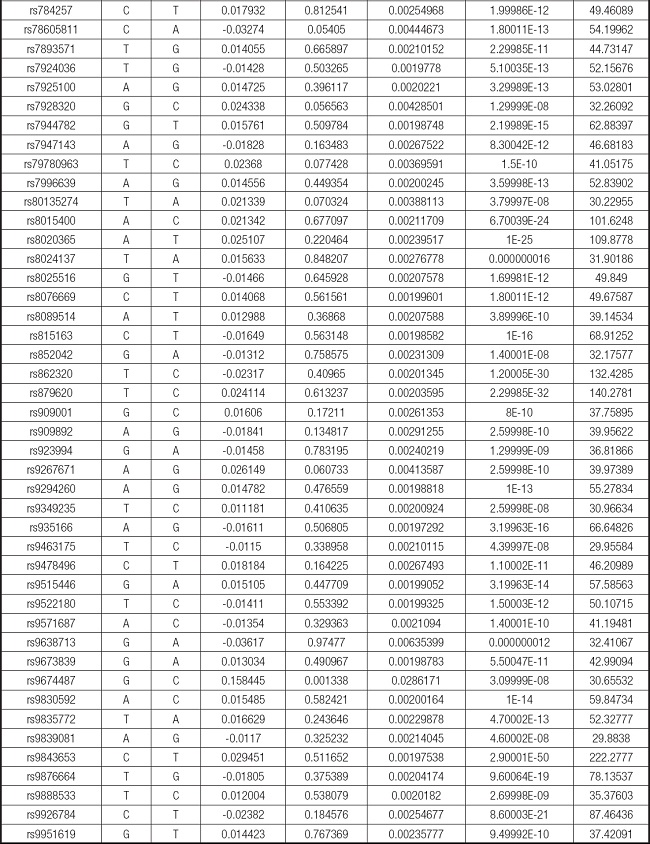
We strictly followed the selection criteria for the genetic instrumental variables. The details of the SNPs finally selected for MR analysis are shown in Supplementary Table I.
THE CAUSAL RELATIONSHIP BETWEEN BMI AND MASTITIS
We evaluated the causal relationship of BMI on mastitis. The results are summarized in figure 3.
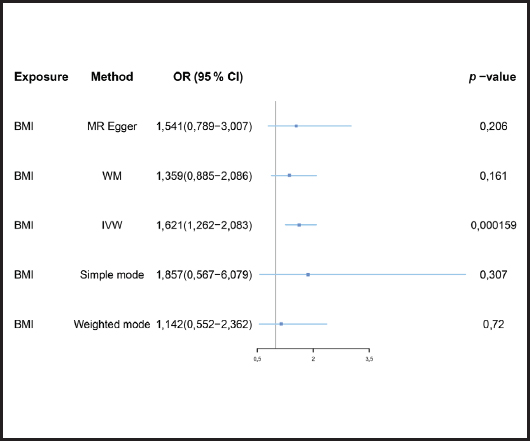
Figure 3. Forest plot of MR analysis results (BMI: body mass index; MR: Mendelian randomization; IVW: inverse-variance weighted; MR-PRESSO: MR-Pleiotropy Residual Sum and Outlier method).
Causal relationships assessed by IVW random effects models indicated that each standard deviation (SD) increased genetically determined BMI increased the risk of mastitis by 62.1 % (n = 413 SNPs, OR = 1.621, 95 % CI: 1.262-2.083, p = 1.59E-4),and this causality was further verified by MR Egger (OR = 1.541, 95 % CI: 0.789-3.007; p = 0.206), weighted median (OR = 1.359, 95 % CI: 0.885-2.086; p = 0.161), simple mode (OR = 1.857,95 % CI: 0.567-6.079; p = 0.307) and weighted mode (OR = 1.142, 95 % CI: 0.552-2.362; p = 0.720) methods. Figure 4 shows a scatter plot of the relationship between BMI and mastitis. Figure 5 shows a forest plot of the causal effect of single nucleotide polymorphisms (SNPs) associated with BMI on mastitis.
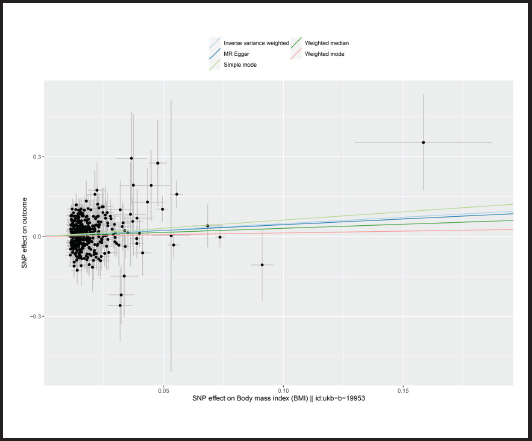
Figure 4. Scatter plot of MR analysis of the effect of BMI on mastitis (BMI: body mass index; SNP: single nucleotide polymorphisms; MR: Mendelian randomization).
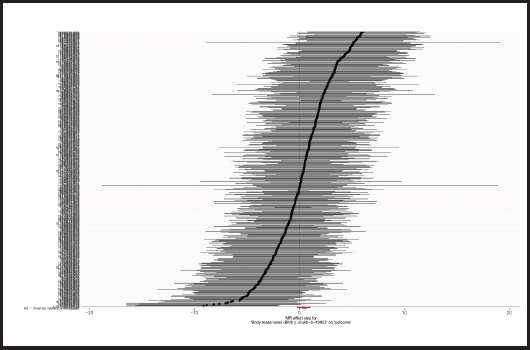
Figure 5. Forest plots of the causal effects of BMI-associated SNPs on mastitis (BMI: body mass index; SNP: single nucleotide polymorphisms; MR: Mendelian randomization).
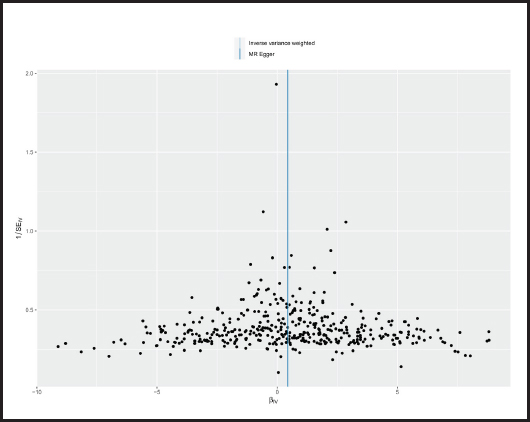
The p-value obtained by Cochran’s Q test was 0.017 < 0.05, so we used the random effects model as the gold standard. According to MR-PRESSO test, no outliers were found, which was consistent with the leave-one-out analysis (Fig. 6) and funnel plot (Fig. 7). Horizontal pleiotropy was absence in MR-Egger intercept analysis (p = 0.873).
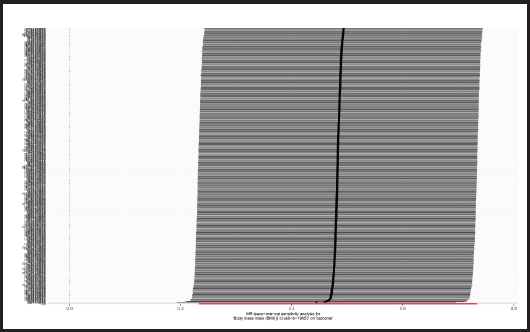
Figure 6. “Leave-one-out” sensitivity analysis results in the two-sample MR analysis (BMI: body mass index; MR: Mendelian randomization).
DISCUSSION
In the present study, we applied MR methods to determine the causal relationship between BMI and mastitis, and we found that for every one standard deviation (SD) increase in BMI, the risk of mastitis increased by 62.1 %.
We selected BMI as the exposed genetic phenotype and found a genetic causal relationship between BMI and mastitis. BMI is often used as a measure of obesity, and the relationship between obesity and mastitis can be reflected laterally through this study (24). Many retrospective studies have been conducted to evaluate potential risk factors for mastitis, however, many of them suffer from recollection and selection bias (25). It has been shown that obesity is a low-grade chronic inflammatory condition with changes in associated factors that can directly affect local estrogen receptor expression and the development of inflammation (26,27). Multiple studies have found that obese individuals are more likely to develop various types of infections than normal weight individuals (28,29). In a previous Mendelian randomization study, high BMI was found to be associated with an increased risk of any infection (30), and infections may contribute to the development of some mastitis. Granulomatous lobular mastitis (GLM) is a benign chronic inflammatory breast disease of unknown etiology, and because GLM is a rare disease, prospective studies are difficult to perform. In a retrospective analysis, obesity or overweight was found to be associated with GLM recurrence (7). Periductal mastitis (PDM) is a long-term inflammatory disease, but little is known about the etiology of PDM. In a retrospective study, overweight/obesity was shown to be an independent risk factor for PDM (6). Non-bacterial mastitis (NBM) is a solid disease of the breast that accounts for 4-5 % of all benign breast diseases (31). In some studies, obesity has been suggested to be possibly related to NBM (32).
There were several advantages for the current two-sample MR studies. First, because alleles follow the principle of random distribution when forming gametes during meiosis, the causal relationship between genotype and disease in MR studies will not be distorted by confounding factors, which is the main limitation of traditional observation studies (33,34). Second, it is much easier to assess the causal relationship between BMI and mastitis risk using the public GWAS database than using prospective cohort studies or randomized controlled trials. This study also required less time and cost than a general epidemiological study.
However, we acknowledge some limitations to our study. First, there might be an ethnic bias in our study due to all the selected GWAS database populations being of European ancestry. Second, there was some heterogeneity in our results in the heterogeneity test, which may have compromised the accuracy of our results, although the results of our other analytical methods were robust. Third, our study did not differentiate between the different types of mastitis, and the relationship between BMI values and each type of mastitis needs further study. Fourth, this study needs to be further validated by other types of studies (observational studies, cohort studies, etc.).
CONCLUSIONS
According to our study, there is a genetic causal relationship between an increased number of BMI and an increased risk of inflammatory breast disease. Patients with high BMI need to control their weight to reduce the incidence of mastitis, and clinicians may also need to be concerned about the potential risk of inflammatory breast disease in some patients with high BMI.