Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
International Microbiology
versión impresa ISSN 1139-6709
INT. MICROBIOL. vol.8 no.1 mar. 2005
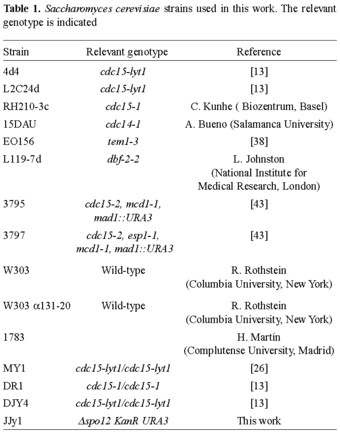
| RESEARCH ARTICLE | |||
|
| |||
| The role of MEN (mitosis exit network) proteins in the cytokinesis of Saccharomyces cerevisiae
Summary. At the latest stages of their cell cycle, cells carry out crucial processes for the correct segregation of their genetic and cytoplasmic material. In this work, we provide evidence demonstrating that the cell cycle arrest of some MEN (mitosis exit network) mutants in the anaphase-telophase transition is bypassed. In addition, the ability of cdc15 diploid mutant strains to develop non-septated chains of cells, supported by nuclear division, is shown. This phenotype is also displayed by haploid cdc15 mutant strains when cell lysis is prevented by osmotic protection, and shared by other MEN mutants. By contrast, anaphase-telophase arrest is strictly observed in double MEN-FEAR (fourteen early anaphase release) mutants. In this context, the overexpression of a FEAR component, SPO12, in a MEN mutant background enhances the ability of MEN mutants to bypass cell cycle arrest. Taken together, these data suggest a critical role of Cdc15 and other MEN proteins in cytokinesis, allowing a new model for their cellular function to be proposed. [Int Microbiol 2005; 8(1):33-42] Key words: Saccharomyces cerevisiae · MEN (mitosis exit network) · Cdc15 · cytokinesis · cell cycle · anaphase-telophase transition · FEAR (fourteen early anaphase release) | ||
| |||
| Función de las proteínas MEN ("mitosis exit network") en la citocinesis de Saccharomyces cerevisiae Resumen. En las últimas etapas de su ciclo celular, las células llevan a cabo procesos cruciales para la segregación correcta del material genético y citoplásmico. Es un tema de investigación de gran actualidad. En este trabajo aportamos pruebas que demuestran que en algunos mutantes MEN ("mitosis exit network") el ciclo celular no se detiene en la transición anafase-telofase. Además, se demuestra la capacidad de las cepas mutantes diploides cdc15 para desarrollar cadenas de células no septadas acompañadas por división nuclear. También muestran ese fenotipo las cepas mutantes haploides cdc15 cuando se impide la lisis celular mediante protección osmótica y lo comparten con otros mutantes MEN. En cambio, la detención en la transición anafase-telofase se observa siempre en los mutantes dobles MEN-FEAR ("fourteen early anaphase release"). En este contexto, la sobreexpresión de un componente FEAR, SPO12, en un fondo MEN mutante aumenta la capacidad de los mutantes MEN para soslayar la detención del ciclo celular. En conjunto, esos datos indican que la proteína Cdc15 y otras proteínas MEN deben de desempeñar un papel crucial en la citocinesis, lo que permite proponer un nuevo modelo de su función en la célula. [Int Microbiol 2005; 8(1):33-42] Palabras clave: Saccharomyces cerevisiae · MEN ("mitosis exit network") · citocinesis · Cdc15 · ciclo celular · transición anafase-telofase · FEAR ("fourteen early anaphase release") | Função das proteínas MEN ("mitosis exit network") na citocinese de Saccharomyces cerevisiae Resumo. Durante as últimas etapas do ciclo celular acontecem processos cruciais para a correta segregação do material genético e citoplasmático. O presente trabalho mostra evidências que provam que a parada do ciclo celular é ignorada por alguns mutantes das proteínas MEN ("mitosis exit network") durante a anáfase-telófase. Além disso, é mostrada também a habilidade de cepas do mutante diplóide cdc15 para desenvolver cadeias de células asseptadas através de divisão nuclear. Este fenótipo é também mostrado por cepas de células mutantes haplóides cdc15 quando a lise das células é impedida através de proteção osmótica, um fenótipo compartilhado por outros mutantes das MEN. Pelo contrário, a parada durante a anáfase-telófase é rigorosamente mantida em mutantes duplos de MEN-FEAR ("fourteen early anaphase release"). Nesse contexto, a sobre-expressão de um dos componentes de FEAR, o SPO12, em um mutante MEN, aumenta a habilidade do mutante para ignorar a parada do ciclo celular. Em conjunto, estes dados sugerem um papel crítico de Cdc15 e outras proteínas MEN na citocinese, permitindo propor um novo modelo para sua função intracelular. [Int Microbiol 2005; 8(1):33-42] Palavras chave: Saccharomyces cerevisiae · MEN ("mitosis exit network") · citocinese · Cdc15 · ciclo celular · transição anáfase-telófase · FEAR ("fourteen early anaphase release") |
Introduction
The latest stages of the cell cycle involve a group of events of singular importance for correct inheritance of cellular genetic material and, subsequently, cell viability. This is a very active and rapidly evolving field of study, exacerbated by the enormous complexity of the mechanisms being investigated (for reviews, see [6,10,41]). In eukaryotic cells, the cell cycle is directed by CDK protein kinases, such as the CDK of Saccharomyces cerevisiae, Cdc28. The activity of Cdc28 is high throughout the M phase of the cell cycle due to the interaction with B-type cyclins (Clbs). However, in order to accomplish the transition from anaphase to telophase, the inactivation of Cdc28 kinase activity is required (for reviews see [24,41]). This inactivation may occur through two different pathways: (i) ubiquitination and later degradation of Clb2 by the APC/C (anaphase-promoting complex/cyclosome) and the proteasome, and (ii) through the activation of Sic1, a Cdc28-Clb2 activity inhibitor [16]. In both cases, the event responsible is the release of the protein phosphatase Cdc14 from the nucleolus [39,46]. Cdc14 dephosphorylates and activates Sic1, Swi5 (a transcriptional activator of Sic1), and Hct1 (an activator of APC/C for Clb2 ubiquitination and proteolysis) [34]. It is thought that the release of Cdc14 from the nucleolus is directed by the activation of a small GTPase from the Ras family called Tem1 [39], which is located at the SPB (spindle pole body). Tem1 migrates into the daughter cell and is only activated by its GEF module (Lte1) when this organelle is close to the daughter cell cortex [3,37], thus providing time for complete separation and migration of the genetic material into the daughter cell. Tem1 is the head of a signaling cascade integrated by MEN (mitosis exit network) proteins [1,17,23]. Among MEN proteins, there are kinases, such as Cdc15, Cdc5, Dbf2/20 [14,15,35]; phosphatases, such as Cdc14 [47]; Tem1, a GTPase as previously mentioned [38]; GTPase regulators, such as Lte1 (GEF of Tem1) [37], Bub2 and Bfa2 (GAP of Tem1) [29,48]; and other proteins with an unclear function in this pathway, such as Spo12 [26,45] and Mbo1 [23].
Recent studies have helped to shed light on several intriguing but unresolved questions. The separation of genetic material, as mentioned above, is crucial for successful cell division. Hence, the regulation of this process must be more delicate than checking SPB positioning by Tem1-Lte1 interaction and activation [33]. In other words, there must be other controls at the level of separation of the genetic material besides those related to SPB localization. Amon et al. [43] identified a new regulatory network, named, FEAR (cdc fourteen early anaphase release), comprising the proteins Cdc5, Esp1, Spo12, and Slk19, which are responsible for a first and early wave of Cdc14 activity in the cell. The model proposed by the authors involves two waves of Cdc14 activity. The first, occurring in early anaphase, is responsible for MEN activation and is induced by FEAR genes. The second and more extensive release of Cdc14 from the nucleolus, at the anaphase-telophase transition, would be determined by the function of the previously activated MEN genes. In addition, these authors and others [30,43] suggested a different spatial distribution of Cdc14: the "early" released Cdc14 molecules could be confined to the nucleus, while the "late" released molecules could reach the cytoplasm, thus having different targets in the different cellular compartments. One of these targets is the GAP module (Bfa1-Bub2) of the above-mentioned Tem1. The consequence of the dephosphorylation and activation of Bfa1 by Cdc14 at the end of telophase is inactivation of the MEN pathway, restoring the initial state prior to the first Cdc14 release [30]. For an exhaustive review of Cdc14, MEN and FEAR see [7]. In switching-off mitosis, recent work has revealed the existence of a mechanism called AMEN (antagonist of the MEN pathway), in which a protein (Amn1) is able to bind directly to Tem1 and prevent association with other MEN proteins [49], thus contributing to MEN pathway inactivation.
MEN proteins have been related to the cytokinesis process. In this context, MEN mutants are unable to initiate and develop septum construction. There is evidence to suggest that MEN proteins are involved in the transmission of the septation signal from the cell cycle machinery to the septum area [8,13,18,20,22,25]. Thus, MEN proteins, through an as yet unclear mechanism, could be responsible for the coordination between cell cycle progression and morphogenesis of the latest stages of the cycle [11]. One of the phenotypic characteristics of the MEN mutants of S. cerevisiae is cell cycle arrest in the anaphase-telophase transition [44]. In previous work by our group, the cell cycle arrest of these mutants was shown to be transient and dependent upon a morphogenetic checkpoint [13]. Thus, when the checkpoint mechanism decays the mutant cells are able to re-bud and continue an anomalous cell cycle and morphogenesis that end with lysis [13].
In this work, we provide evidence that cdc15 and other MEN mutations do not definitively arrest the cell cycle and we propose a new model for the function of MEN genes in the late stages of mitosis.
Materials and methods
Strains, media, and culture conditions. The Escherichia coli strain for DNA manipulations was DH5α. Molecular manipulations were carried out as described in [32]. The yeast strains and their genotypes are listed in Table 1. For observation and quantification of chained cells, strains were grown at 24ºC for 16 h in a flask containing 100 ml yeast peptone dextrose (YPD) medium. Optical density (600 nm) was adjusted to 0.1 by refreshing with YPD followed by incubation at 37ºC. Aliquots were taken at the indicated times and the phenotype was observed under a microscope as detailed below. In experiments in which osmotic protection was required, 1 M sorbitol or 0.2 M NaCl were added to the YPD at both the growth and incubation stages. In assays in which Clb2 was overproduced, the strains were grown in SD media containing 2% glucose and then shifted to SD medium containing 2% galactose and raffinose, instead of glucose, as the only carbon source.
SPO12 cloning and deletion. SPO12 was cloned by PCR using the following oligonucleotides: 5'-GTCGACGATTCCCATTGTATTGCCTC-3' (upper) and 5'-GTCGACATCAAGGTTTCATAATTTGGA-3' (lower); underlined regions are artificial SalI sites introduced to facilitate molecular manipulations. A 1-kb fragment (containing the ORF, 360 bp upstream from the ATG, and 176 downstream from the stop codon) was amplified, sequenced to detect any PCR mistakes, and cloned into the pGEMT cloning vector (Promega). The pGEMt-SPO12 plasmid was SalI-digested and the 1-kb fragment obtained was introduced into the YEp352 and YCplac33 vectors, affording YEpSPO12 and YCpSPO12 plasmids, respectively.
To delete the full sequence of SPO12 ORF, the PCR method described in [21] was used. The following oligonuclotides were designed:
5'-CAAAATAACATATACAGTAAGAACAATAGAAAACGTATTTCGGATCCCCGGGTTAATTAA-3' (upper) and 5'-GTAGCATTTGGCTATTTTTGGATGACTAGAAAGGCAGATTGAATTCGAGCTCGTTTAAAC-3' (lower); underlined regions are the SPO12-specific sequences. A 600-bp fragment was obtained by PCR using as template plasmid pFA6-MX4, in which the genetic marker was kanamycin resistance. The amounts of DNA resulting from ten PCR reactions were pooled, precipitated, resuspended in 5 µl of water and transformed into the S. cerevisiae strain 1783 by the lithium acetate method [32]. The transformants were selected on YPD plates containing G418 at a final concentration of 200 µg/ml. Correct deletion of the SPO12 ORF was checked by PCR, using as template the genomic DNA from the transformants and as primers the lower oligonucleotide, designed in the kanamycin gene (5'-CAGCGTGGACTAACGGGCTGT-3' ), and the same upper primer used for cloning the gene as described above. The deleted strain thus generated was called JJy1. Attempts to delete SPO12 in a cdc15-lyt1 background (the strain 4d4) were unsuccessful (see above).
Regulation of Clb2 expression. The diploid mutant strain DJY4 (cdc15-lyt1/cdc15-lyt1) was transformed with the plasmid (GAL-CLB2) containing the Clb2 cyclin under control of the galactose promoter, thereby overproducing Clb2 when the only carbon source was galactose. SPO12 was introduced into an episomic plasmid and into a centromeric plasmid (YEp352 and YCplac33, respectively). The two plasmids, called YEpSPO12 and YCpSPO12, were transformed into the following MEN mutant strains: 4d4 (cdc15-lyt1), EO156 (tem1-3), LH119-7d (dbf2), and 15DAU (cdc14-1).
Staining procedures and microscopy. Nucleus staining was carried out using the DAPI method. From a cell suspension, 3 µl was spread out on a microscope slide and a DAPI solution (2.5 µg/ml) was added as described [31]. Cells were observed under a Leica (model DMRXA) fluorescence microscope. Images were recorded with a camera from Photometrix Sensys run with the Q-fish program and were processed using Adobe Photosho
Results
Diploid strains defective in CDC15 bypass the anaphase-telophase cell cycle arrest. When incubated at the restrictive temperature, cdc15 diploid mutant strains developed chains of non-septated cells, each of them containing a nucleus, as observed by DAPI staining (Fig. 1) [13]. This feature was independent of the cdc15 mutant alleles carried by the diploid mutant strain; thus, similar results were obtained with different diploid strains bearing different cdc15 mutant alleles, such as DR1 (cdc15-1/cdc15-1), DJY4 (cdc15-lyt1/cdc15-lyt1), and MY1 (cdc15-1/cdc15-lyt1)[13]. Furthermore, the amount of DNA of the different cdc15 diploid mutant strains was measured by flow cytometry, and a population of cells with increased amounts of DNA was obtained (data not shown), in consonance with the above-reported nuclear staining findings.
Overexpression of Clb2 arrests the cell cycle in the anaphase-telophase transition, probably through saturation of the degradation machinery [44]. Clb2 overexpression in strain DJY4 incubated at the restrictive temperature produced a sharp decrease in the percentage of chained cells (12%) compared with the control strain displaying normal levels of the cyclin (37%). These results suggest normal functioning of the cell cycle machinery in cdc15 mutants.
Haploid strains defective in cdc15, kept alive by osmotic protection, bypass cell cycle arrest. In haploid cdc15 mutant strains, prosecution of cell morphogenesis and the cell cycle occurred; however, these were accompanied by the formation of abnormal apical projections corresponding to a new (and anomalous in terms of polarity and morphogenesis) bud process that ended with cell lysis. The intensity, but not the presence, of this phenomenon was dependent on the background and culture conditions, such as refreshing the culture medium immediately before incubation at the restrictive temperature (data not shown).
In order to maintain the cells ability to bypass cell cycle arrest but remain alive, i.e. avoiding cell lysis, and hence to allow the evolution of the cell cycle in these haploid strains to be studied, an osmotic stabilizer was added. Under these new conditions, a significant percentage of haploid cells developed a chained-cell phenotype identical to that shown by the diploid mutant strains described previously in this work (Fig. 2). No chained cells were observed when the cultures were incubated in the absence of osmotic protection. In order to rule out possible effects of 1 M sorbitol other than osmotic protection, another osmotic stabilizer (0.2 N NaCl; osmotic strength comparable to 1 M sorbitol) was tested and the results were identical to those obtained when 1 M sorbitol was used (data not shown). These observations indicate that, when cell lysis is prevented in haploid cdc15 mutant strains, their cell cycle is able to continue in the absence of septation, rendering the above-described chained-cell phenotype.
Behavior of other MEN mutants. When cdc14-1, dbf2-2, and tem1-3 mutant strains were incubated at the restrictive temperature and cell lysis was prevented by adding 1 M sorbitol, all the strains developed a phenotype of chained cells like that reported above for cdc15 mutants (Fig. 2). As in the cdc15 mutant strain, no chained cells were seen when the experiments were carried out in the absence of sorbitol. Thus, to some extent, all of the strains were able to bypass the anaphase-telophase transition arrest and continue the cell cycle upon incubation at the restrictive temperature and in the presence of osmotic stabilization. Figure 2 shows only the chained cells, but it is interesting to note that almost all the non-scored cells bypassed the cell cycle arrest and developed the above-described aberrant morphology, leading to cell lysis. It should also be noted that no chained cells were found in these mutant strains when they were grown at the permissive temperature.
A double mutant in a component of MEN and in a component of FEAR. Previous results in this work had shown that the cell cycle is not arrested in the absence of MEN proteins. The role of MEN in the release of Cdc14 from the nucleolus and, through the activation of Hct1 and Sic1, in the elimination of Clb2 and CDK inactivation must be recalled. Thus, the above results suggest the existence of a parallel mechanism for CDK-cyclin inactivation in the anaphase-telophase transition other than the inactivation determined by Sic1 or Hct1 and, for instance, MEN. In this context, during this work, the existence of a mechanism-called FEAR-responsible for an early and slight release of Cdc14 from the nucleolus was reported [43]. In light of this, we examined whether the early and very small release of Cdc14 due to FEAR might be that parallel mechanism for the cell cycle to continue in MEN mutants.
Strain 3797(cdc15-2, esp1-1), which is a double mutant bearing a mutation in a MEN component and in a FEAR component [43], could not form chained cells when the microorganism was incubated at the restrictive temperature.. This confirmed the cell cycle arrest at the anaphase-telophase transition (Fig. 3). As controls of the experiment, the isogenic strain of strain 3797 (bearing the cdc15-2 mutation) and the double-mutant strain 3797 transformed with a plasmid containing the CDC15 wild-type gene were used. As expected, in the first control strain (bearing the cdc15 mutation), there was a significant proportion of chained cells, while in the second control strain (bearing the esp1 mutation) chained cells were undetectable. When the experiment was carried out in the presence of osmotic stabilization, a higher proportion of chained cells was observed in the case of the strain bearing the single cdc15 mutation while no chained cells were observed for the double mutant cdc15, esp1 strain (Fig. 3).
To expand our data concerning the above MEN-FEAR, we attempted to generate another double MEN-FEAR mutant: namely, a cdc15-lyt1, Δspo12 double-mutant strain. SPO12 was deleted in a cdc15-lyt1 background using a PCR-based method, and the appropriate mutant strains (4d4, a cdc15-lyt1 strain and JJy1, a Δspo12 strain) were mated. However, neither of the methods afforded the double-mutant strain. Statistically, the high number of tetrads analyzed by the genetic method confirms the impossibility of the coexistence of both mutations. The same conclusion was drawn in another study [26] and was explained in terms of the fact that spores bearing the double mutation were unable to germinate.
Overproduction of Spo12 in a MEN background. The sequence of the cloned SPO12 ORF showed no differences with the sequence deposited in the databases. Spo12 was transformed into different MEN mutants, either in an episomic or a centromeric plasmid. After incubation of the transformed strains at 37ºC, there was a significant increase in the proportion of chained cells, which correlated with the level of Spo12 expression in the cdc15-lyt1 and tem1-3 mutants (Fig. 4A, B); the dbf2 mutation was suppressed (Fig. 4C), as previously described [28], and the cdc14-1 mutant was unaffected (Fig. 4D).
Discussion
What are chained cells and what do they mean? The diploid strains bearing a cdc15 mutation had a remarkable proportion of chained-unseptated cells with a nucleus in each cell. This phenotypic trait points to cell cycle progression in the absence of septation. Chained-cell formation is a typical feature of mutants affected in cytokinesis but not in cell cycle progression, such as myo1 [19] or Chs2 [36]. We therefore decided to use the chained-cell phenotype as an indicator of the bypassing in late mitosis of cell cycle arrest in the absence of cytokinesis. This tool allowed us to study the role played by MEN proteins in the latest stages of the cell cycle. Interestingly, the proportion of chained cells in haploid strains was practically negligible, and, instead of chained cells enlarged cells bearing an aberrant (in terms of cell polarity) apical projection, corresponding to a new budding process., were found The fate of these mutants is cell lysis. This phenotype has been described in depth and is explained in [13].
Why are haploid strains unable to develop chained cells? The answer could lie in the different polarity patterns of haploid and diploid strains [5]. Logically, a haploid cell budding in a bipolar fashion violates its intrinsic pattern of polarity and must be unable to correctly build all the structures needed and hence, finally, cell lysis occurs. The opposite arises in diploids because there is no change in the polarity pattern; hence, the architecture of the cells must be more stable and able to support growth in non-septated chains of cells. In this context, and reinforcing this idea, when a haploid strain was protected from lysis by an iso-osmotic culture medium, the haploid cells were able to develop the chained-cells phenotype.
The phenotype of cdc15 and other MEN mutants. Classically, MEN proteins have been implicated in cell cycle progression [12,27,44,50]. This implies that MEN mutants must cell cycle arrest in the anaphase-telophase transition. Data indicating the transient character of cell cycle arrest in MEN mutants as result of the occurrence of a checkpoint have been reported [13]. In agreement with those previous results, the phenotypic data presented here demonstrate the bypassing of cell cycle arrest in the anaphase-telophase transition by MEN mutants. Additionally, these mutants are affected in cytokinesis and are completely unable to carry out the septation process [13]. Also, using different approaches, several authors have described the relationship between MEN proteins and cytokinesis, including coordination between cytokinesis and nuclear separation [8,11,18,20,22,25,42]. To explain our initial observations (cell cycle prosecution in MEN mutants), two different hypotheses were initially considered: (i) A parallel mechanism responsible for CDK-Clb2 complex inactivation, and thus responsible for the cell cycle progression observed here, that differs from processes regulated by MEN proteins (Sic1 and Hct1) must exist. (ii) Regulation of cell cycle progression is not driven directly by MEN proteins, and their role is primarily related to the septation process and to the coordination between the cell cycle and morphogenesis.
Recently, new clues regarding the regulation of the anaphase-telophase transition have been provided. This new mechanism, called FEAR [30,43], could be the hypothetical parallel mechanism proposed above. To investigate this possibility, the cell cycle-arrest phenotype of a double mutant in both mechanisms, MEN and FEAR; namely, a cdc15, esp1 double mutant, was analyzed. (The genotype of this mutant is more complex such that the cell cycle arrest driven by the SPB checkpoint mechanism due to the esp1 mutation will not occur, as described in detail by Stegmeier [43].) This double mutant did not show chained cells, and hence the cell cycle must strictly arrest at the anaphase-telophase transition. Another putative parallel mechanism for CDK-Clb2 inactivation different from Hct1 and Sic1 has been described [4] in which Cdc6, a protein with structural similarity to Sic1, is involved in the inactivation of the CDK. Nevertheless, the structural similarity with the components of the mechanisms governed by MEN proteins, as well as preliminary data (not shown), reduces interest in Cdc6 as the hypothetical parallel inactivation mechanism.
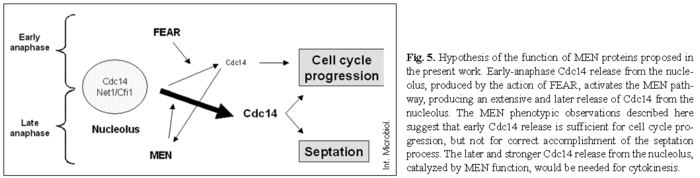
A new model for the action of MEN proteins. The new data discussed above, together with our results, allow us to propose a new model for the function of the MEN and FEAR groups of proteins. This model could be considered a combination of the two earlier hypotheses. FEAR proteins could indeed be the parallel mechanism for CDK-Clb2 inactivation in the absence of MEN proteins, and MEN proteins could be essential for septum morphogenesis, in addition to their cell cycle role. According to the generally accepted model, FEAR proteins are responsible for the early and slight release of Cdc14 from the nucleolus, as described in [30,43]. This early released Cdc14 activates the MEN mechanism [30], which is responsible for late and extensive release of Cdc14 from the nucleolus [30,43].
Our contribution to the present model is that early Cdc14 release seems to be sufficient for cell cycle progression (even in the absence of MEN); but the late and extensive release, induced by MEN proteins, is required for the septation process (Fig. 5). This model is supported by the absence of chained cells when a FEAR component was absent in a MEN mutant strain, and by the observation of chained cells when a FEAR protein was ectopically overproduced in a MEN mutant background. The increased level of a FEAR protein would be able to induce the bypassing of cell cycle arrest owing to the induction of early Cdc14 release, affording chained cells; but it would be unable to induce late Cdc14 release, preventing cytokinesis from occurring, and thus the appearance of chained cells.
This model implies that the targets of Cdc14 in each early and late wave must be different, most likely due to a different spatial localization of Cdc14, as proposed by Schiebel's group[30]. Currently, this is a very exciting but not fully understood field of study. Apart from the classical localization, dynamics, and targets of Cdc14 [39,46], there are reports of Cdc14 localized at the SPB [30,51], where it would regulate the activity of MEN proteins by direct interaction with different components, such us Bfa1 and Tem1[30] and Cdc15[43]. (For an excellent review of this topic, see [9].)
The new model proposed here supports the central role played by MEN proteins in morphogenesis at the septum area, and provides additional evidence of the critical function of Cdc14 in both processes: the cell cycle and morphogenesis. This model is in concordance with the findings of other authors, who have proposed that the main role of MEN proteins is the release of Cdc14 from the nucleolus [40]. In addition to our findings, new and interesting evidence exists, although not fully formulated, to suggest a role of MEN in the regulation of nuclear transport [2,40]. As a result, it is tempting to speculate that MEN proteins, in some unknown way, are responsible for the exit of late-released Cdc14 from the nucleus, allowing its interaction with targets related to septum formation. Nevertheless, this notion is fraught with unresolved questions, such as the regulation of nuclear transit by proteins localized at the SPB.
Are Dbf2 and Cdc14 different from the other MEN proteins? So far, in the discussion of this work, it has been assumed that all MEN proteins are involved in the same processes. Nevertheless, we several phenotypic traits exclusive to the dbf2 mutant strain and not shared by other MEN mutants have been addressed. First, the proportion of chained cells was the lowest among the MEN mutants (Fig. 2) and, second, when Spo12 was overproduced in a dbf2 mutant strain both septation and cell cycle defects in the mutant were suppressed (Fig. 4C) [28]. It could therefore be envisaged that the role played by Dbf2 must somehow be different from those played by the rest of the MEN mutants and must be related both to cytokinesis and cell cycle progression. It should be recalled that Dbf2 is a MEN protein that is able to move from the SPB to the neck, and it is thought to be the key for linking cell cycle signals and septum morphogenesis [11]. Another alternative explanation has been suggested in [28]; the authors proposed a direct interaction between these proteins and suggested that Spo12 regulates the function of Dbf2.
It is common for the Cdc14 to be included as an MEN protein, but it should be noted that, although similar to MEN proteins, Cdc14 and its function do in fact differ from them with respect to subcellular localization and the central role played by Cdc14 in cell cycle regulation. It is clear that Cdc14 is hierarchically more important for the cell cycle than the other MEN proteins. This could explain why cdc14 mutants were unaffected by the overproduction of Spo12, in contrast to the phenotype of the rest of the MEN proteins.
Concluding remarks. Our findings suggest that the action of FEAR proteins is to enhance cell cycle progression in some MEN mutant backgrounds (cdc15 and tem1), but that they are not be related to the cytokinesis process, with the exception of the dbf2 mutant background, in which both processes, cell cycle progression and cytokinesis, are affected. The main issue remaining is: why a cdc14-1 mutant allele is able to bypass the anaphase-telophase cell cycle arrest but is unable to septate Our guess would be that this thermosensitive allele has sufficient activity to promote cell cycle prosecution but not cytokinesis, either through some remnant activity or through a modular affectation of the function. The functional differences among MEN proteins challenge us to question their involvement in several mechanisms that are the responsibility of different-and individual-components of the pathway, suggesting a higher degree of complexity than is currently known.
Acknowledgements. We would like to acknowledge to C. Kunhe, A. Toh-e, A. Amon, and A. Bueno for their yeast strains; and M Glozer for the GAL-CLB2 overexpression plasmid. We also thank Víctor J. Cid and Jaime Correa for critical reading of the manuscript. This work was made possible by a FEDER project from the European Union (IFD97-1897-CO2-01).
References
1. Asakawa K, Yoshida S, Otake F, Toh-e A (2001) A novel functional domain of Cdc15 kinase is required for its interaction with Tem1 GTPase in Saccharomyces cerevisiae. Genetics 157:1437-1450 [ Links ]
2. Asakawa K, Toh-e A (2002) A defect of Kap104 alleviates the requirement of mitotic exit network gene functions in Saccharomyces cerevisiae. Genetics 162:1545-1556 [ Links ]
3. Bardin AJ, Visintin R, Amon A (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell 102:21-31 [ Links ]
4. Calzada A, Sacristan M, Sanchez E, Bueno A (2001) Cdc6 cooperates with Sic1 and Hct1 to inactivate mitotic cyclin-dependent kinases. Nature 412:355-358 [ Links ]
5. Chant J, Pringle JR (1995) Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol 129:751-765 [ Links ]
6. Cid VJ, Jimenez J, Molina M, Sanchez M, Nombela C, Thorner JW (2002) Orchestrating the cell cycle in yeast: sequential localization of key mitotic regulators at the spindle pole and the bud neck. Microbiology 148:2647-2659 [ Links ]
7. D'Amours D, Amon A (2004) At the interface between signaling and executing anaphase-Cdc14 and the FEAR network. Genes Dev 18:2581-2595 [ Links ]
8. Frenz LM, Lee SE, Fesquet D, Johnston LH (2000) The budding yeast Dbf2 protein kinase localises to the centrosome and moves to the bud neck in late mitosis. J Cell Sci 113:3399-3408 [ Links ]
9. Geymonat M, Jensen S, Johnston L (2002) Mitotic exit: the Cdc14 double cross. Curr Biol 12:R482-484 [ Links ]
10. Guertin DA, Trautmann S, McCollum D (2002) Cytokinesis in eukaryotes. Microbiol Mol Biol Rev 66:155-178 [ Links ]
11. Hwa Lim H, Yeong FM, Surana U (2003) Inactivation of mitotic kinase triggers translocation of MEN components to mother-daughter neck in yeast. Mol Biol Cell 14:4734-4743 [ Links ]
12. Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO (1998) A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell 9:2803-2817 [ Links ]
13. Jimenez J, Cid VJ, Cenamor R, Yuste M, Molero G, Nombela C, Sanchez M (1998) Morphogenesis beyond cytokinetic arrest in Saccharomyces cerevisiae. J Cell Biol 143:1617-1634 [ Links ]
14. Johnston LH, Eberly SL, Chapman JW, Araki H, Sugino A (1990) The product of the Saccharomyces cerevisiae cell cycle gene DBF2 has homology with protein kinases and is periodically expressed in the cell cycle. Mol Cell Biol 10:1358-1366 [ Links ]
15. Kitada K, Johnson AL, Johnston LH, Sugino A (1993) A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol 13:4445-4457 [ Links ]
16. Knapp D, Bhoite L, Stillman DJ, Nasmyth K (1996) The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1. Mol Cell Biol 16:5701-5707 [ Links ]
17. Lee SE, Frenz LM, Wells NJ, Johnson AL, Johnston LH (2001) Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr Biol 11:784-788 [ Links ]
18. Lee SE, Jensen S, Frenz LM, Johnson AL, Fesquet D, Johnston LH (2001) The Bub2-dependent mitotic pathway in yeast acts every cell cycle and regulates cytokinesis. J Cell Sci 114:2345-2354 [ Links ]
19. Lippincott J, Li R (1998) Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol 140:355-366 [ Links ]
20. Lippincott J, Shannon KB, Shou W, Deshaies RJ, Li R (2001) The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J Cell Sci 114:1379-1386 [ Links ]
21. Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961 [ Links ]
22. Luca FC, Mody M, Kurischko C, Roof DM, Giddings TH, Winey M (2001) Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol Cell Biol 21:6972-6983 [ Links ]
23. Mah AS, Jang J, Deshaies RJ (2001) Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc Natl Acad Sci USA 98:7325-7330 [ Links ]
24. McCollum D, Gould KL, Asleson CM, Bensen ES, Gale CA, Melms AS, Kurischko C, Berman J (2001) Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol 11:89-95 [ Links ]
25. Menssen R, Neutzner A, Seufert W (2001) Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr Biol 11:345-350 [ Links ]
26. Molero G, Yuste-Rojas M, Montesi A, Vazquez A, Nombela C, Sanchez M (1993) A cdc-like autolytic Saccharomyces cerevisiae mutant altered in budding site selection is complemented by SPO12, a sporulation gene. J Bacteriol 175:6562-6570 [ Links ]
27. Morgan DO (1999) Regulation of the APC and the exit from mitosis. Nat Cell Biol 1:47-53 [ Links ]
28. Parkes V, Johnston LH (1992) SPO12 and SIT4 suppress mutations in DBF2, which encodes a cell cycle protein kinase that is periodically expressed. Nucleic Acids Res 20:5617-5623 [ Links ]
29. Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell 6:1-10 [ Links ]
30. Pereira G, Manson C, Grindlay J, Schiebel E (2002) Regulation of the Bfa1p-Bub2p complex at spindle pole bodies by the cell cycle phosphatase Cdc14p. J Cell Biol 157:367-379 [ Links ]
31. Pringle JR, Adams AE, Drubin DG, Haarer BK (1991) Immunofluorescence methods for yeast. Methods Enzymol 194:565-602 [ Links ]
32. Sambrook J, Russell DW (2001) Molecular cloning, a practical approach. 3rd edn. Cold Spring Harbor Laboratory Press, New York [ Links ]
33. Saunders WS (2002) The FEAR factor. Mol Cell 9:207-209 [ Links ]
34. Schwab M, Lutum AS, Seufert W (1997) Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell 90:683-693 [ Links ]
35. Schweitzer B, Philippsen P (1991) CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast 7:265-273 [ Links ]
36. Shaw JA, Mol PC, Bowers B, Silverman SJ, Valdivieso MH, Duran A, Cabib E (1991) The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J Cell Biol 114:111-123 [ Links ]
37. Shirayama M, Matsui Y, Tanaka K, Toh-e A (1994) Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast 10:451-461 [ Links ]
38. Shirayama M, Matsui Y, Toh-e A (1994) The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol 14:7476-7482 [ Links ]
39. Shou W et al. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244 [ Links ]
40. Shou W, Deshaies RJ (2002) Multiple telophase arrest bypassed (tab) mutants alleviate the essential requirement for Cdc15 in exit from mitosis in S. cerevisiae. BMC Genet 3:4 [available at http://www.biomedcentral.com/1471-2156/3/4] [ Links ]
41. Simanis V (2003) The mitotic exit and septation initiation networks. J Cell Sci 116: 4261-4262 [ Links ]
42. Song S, Lee KS (2001) A novel function of Saccharomyces cerevisiae CDC5 in cytokinesis. J Cell Biol 152:451-469 [ Links ]
43. Stegmeier F, Visintin R, Amon A (2002) Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell 108:207-220 [ Links ]
44. Surana U, Amon A, Dowzer C, McGrew J, Byers B, Nasmyth K (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J 12:1969-1978 [ Links ]
45. Toyn JH, Johnston LH (1993) Spo12 is a limiting factor that interacts with the cell cycle protein kinases Dbf2 and Dbf20, which are involved in mitotic chromatid disjunction. Genetics 135:963-971 [ Links ]
46. Visintin R, Hwang ES, Amon A (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature 398:818-823 [ Links ]
47. Wan J, Xu H, Grunstein M (1992) CDC14 of Saccharomyces cerevisiae. Cloning, sequence analysis, and transcription during the cell cycle. J Biol Chem 267:11274-11280 [ Links ]
48. Wang Y, Hu F, Elledge SJ (2000) The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol 10:1379-1382 [ Links ]
49. Wang Y, Shirogane T, Liu D, Harper JW, Elledge SJ (2003) Exit from exit. Resetting the cell cycle through Amn1 inhibition of G protein signaling. Cell 112:697-709 [ Links ]
50. Wasch R, Cross FR (2002) APC-dependent proteolysis of the mitotic cyclin Clb2 is essential for mitotic exit. Nature 418:556-562 [ Links ]
51. Yoshida S, Asakawa K, Toh-e A (2002) Mitotic exit network controls the localization of Cdc14 to the spindle pole body in Saccharomyces cerevisiae. Curr Biol 12:944-950 [ Links ]