Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Revista de Osteoporosis y Metabolismo Mineral
versión On-line ISSN 2173-2345versión impresa ISSN 1889-836X
Rev Osteoporos Metab Miner vol.12 no.3 Madrid jul./sep. 2020 Epub 25-Ene-2021
https://dx.doi.org/10.4321/s1889-836x2020000300005
ORIGINALS
Study of bone factor expression in murine model in the absence of pleiotrophin and its changes in the inflammatory situation
1Institute of Applied Medicine. Department of Basic Medical Sciences. Faculty of Medicine
2Department of Chemistry and Biochemistry. Faculty of Pharmacy
3Department of Pharmaceutical and Health Sciences. Faculty of Pharmacy
Pleiotrophin (PTN) is a peptide involved in the development and maintenance of bone tissue with important functions in inflammatory processes. However, the deletion of PTN in murine models does not produce a significant bone deterioration, but the mechanisms that compensate for its loss have not been studied to date. Our study was aimed at verifying how the deletion of PTN and acute inflammation affect the expression of bone factors. To this end, we used three-month-old female mice deficient for PTN (PTNKo) to which we induced acute inflammation by administration of lipopolysaccharide (LPS). Vertebrae and tibiae were isolated to measure gene expression and carry out an osteocyte count. In cell cultures, we checked whether PTN could protect MC3T3 (osteoblast) and MLOY4 (osteocyte) cells from the induction of cell death caused by etoposide. Our results show that the expression of osteocalcin is increased in the vertebrae of PTNKo mice, and that inflammation increased the expression of podhalanin (E11), connexin 43 (Cox43) and the parathormone-related peptide (PTHrP) in the PTNKo mice treated with LPS. Administering PTN significantly reduced etoposide-induced death in MC3T3 and MLOY4 cell cultures. Thus, PTN deficiency induced increased expression of OCN, and acute inflammation produced overexpression of E11, PTHrP, and Cox43 in PTNKo mice. PTN increased the viability of osteoblastic cells and osteocytes compared to etoposide treatment.
Key words pleiotrophin; bone homeostasis; murine model
INTRODUCTION
Pleitropin (PTN) is a cytokine secreted by multiple tissues during embryonic development, and which in adulthood is abundantly expressed in the brain and bone1,2. PTN is composed of 136 amino acids and its sequence is very rich in lysine and cysteine. Together with midkine (Mdk), with which it shares 50% homology, this cytokine constitutes the heparin-binding family of growth and differentiation factors, both having affinity for bone extracellular matrix3-5. PTN is also known as osteoblast stimulating factor 1 (OSF-1) or heparin-linked growth factor (HB-GAM)6. This cytokine was initially isolated from the bone and neuronal tissues of newborn rats2,7,8 and subsequently its homologues have been found in many species including humans, with 90% homology between the different species9,10.
PTN reportedly exerts its effects through its binding to glucosamin-glucans of several receptors such as Nsyndecan, also called syndecan 311, syndecans 1 and 412, integrin αvβ313 and the receptor protein tyrosine phosphatase beta/zeta (PTRPβ/ζ)14. It has also been suggested that nucleolin may be a low-affinity receptor for PTN15 and that anaplastic lymphoma kinase (ALK) may play a role in PTN signaling16.
Probably the best-studied receptor for PTN is PTRPβ/ζ that has also been shown to be expressed in osteoblasts17. This receptor is a protein tyrosine phosphatase whose activation by PTN produces its destabilization and, therefore, the cessation of its phosphatase activity. This triggers the increased phosphorylation of its substrates (e.g. fyn kinase) leading, among other effects, to the activation of the nuclear factor that enhances the kappa light chains of activated B cells (NFκβ)18. The presence of this receptor in bone tissue and its activation through PTN and another ligand, such as insulin-like growth factor binding protein 2 (IGFBP-2)19 has recently been confirmed. Conversely, the expression of N-syndecane in osteoblasts and its relationship with bone regeneration have also been demonstrated6. In this case, the binding of PTN to N-syndecane would produce the phosphorylation of src, which in turn would lead to reorganizations in the cell cytoskeleton that would allow an increase in cell migration20.
Among PTN’s most prominent functions is its role as a promoter of angiogenesis and endothelial cell migration13,21, the growth of neurites22 and its role as a modulator of inflammatory processes governed by microglia in the central nervous system23. The functions of PTN in bone tissue are diverse. Thus, PTN has been described as capable of inducing the proliferation of osteblastic cells in a manner dependent on its concentration and the expression of its receptors24. It promotes the differentiation of mesenchymal cells to chondrocytes during bone development25 and increases migration and adhesion to the extracellular matrix of bone cells6,26. In fact, the role of PTN in relation to bone mass has been investigated by carrying out gene deletion or overexpression experiments in murine models. In the first case, the absence of the gene reportedly did not produce a decrease in bone mass or significant changes in the biomechanical properties of these bones27. Later studies also found that the bone structure of mice without PTN was not altered, but that there was a delay in bone maturation in 2-monthold mice28.
The role of PTN in osteocyte mechanotransduction has also been studied with different results. In in vitro studies, mechanical loading was found to lead to a decrease in PTN expression in SaOs-2 bone cells and in primary osteoblasts subjected to this stimulus29. However, Imai et al. demonstrated that MLOY-4 cells (osteocyte cell line), treated with mechanical loading, increased their production of PTN28. These data are consistent with those found in an animal model of female C57BL/6J mice subjected to mechanical loading, in which there was an increase in the expression of this cytokine29. Furthermore, in the same study, the absence of PTN (using a mouse with the deleted PTN gene) did not influence the increase in bone mass produced by mechanical loading.
Additionally, the effects of overexpression of PTN in bone tissue do seem to have a protective effect in situations of loss of bone mass, such as weightlessness. Mice transgenic for PTN are partially protected against the loss of mass produced by the state of weightlessness when subjected to a stay in the international space station, this protection being related to an increase in osteoblastic activity30.
As has been commented, the absence of PTN in bone does not seem to significantly influence the skeletal structure of murine models, in this work we endeavored to ascertain how the expression of factors associated with the correct maintenance of bone metabolism was altered in the absence of PTN. Furthermore, we wanted to investigate how the expression of these genes was regulated in a situation of acute inflammation produced by injection of LPS, both in the presence and in the absence of PTN. Finally, we verify the protective effect of PTN on osteoblasts and osteocytes in the presence of a death stimulus such as etoposide.
MATERIAL AND METHODS
Animal model
Three-month-old female mice deficient in PTN (PTNKo) and with normal genotype (WT) were used, 9 mice for WT and 9 for PTNKo per group. All the mice came from our animal facility, where they are routinely raised. To induce a state of acute inflammation, a dose of 7.5 mg/kg of lipopolysaccharide was injected intraperitoneally 16 hours before sacrifice in 6 WT mice and 5 PTNKo mice. The animal protocols were approved by the Animal Welfare Committee of the CEU-San Pablo University and the Ministry of the Environment of the Community of Madrid, in accordance with Royal Decree R.D. 53/2013 and European guideline 2010/63/EU.
Processing of bone samples and RNA purification
Total RNA was extracted from the vertebrae after removing non-bony tissues from the lumbar vertebrae 1 to 3, by means of a tissue spray and by dissolving this spray in trizol (Invitrogen, Groningen, The Netherlands). RNA was extracted by means of purifications by the chloroform: isoamyl method (Sigma-Aldrich), followed by precipitation with isopropanol, subsequent washes with 70% ethanol and resuspension in sterile RNAase-free water. RNA reverse transcription to obtain complementary DNA was carried out from 2 µg of RNA using the cDNA High capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, California, USA) in an Eppendorf mastercycler thermocycler, following the following sequential protocol: 10 minutes at 25ºC, 120 minutes at 37º C and 5 minutes at 85ºC.
Quantitative PCR
Quantitative PCR was carried out in an AB 7500 HD thermal cycler (Applied Biosystems). Using TaqMan MGB probes (Assay-by-DesignTM System, Applied Biosystems) to measure the expression of the following genes: osteoprotegerin (OPG), activator receptor ligand for nuclear factor κB (RANKL), osteocalcin (OCN), related peptide parathyroid hormone (PTHrP), podoplanin (E11), connexin 43 (Cox43), vascular endothelial growth factor receptor 2 (VEGFR2) and dickkopf-1 (Dkk1). To produce the PCR reaction, the polymerase included in the Taqman mastermix kit (Applied Biosystems) was used applying the following protocol of 1 minute at 95ºC, followed by 40 cycles of 15 seconds at 95ºC and 1 minute at 60ºC, collecting the fluorescence data in each step of 60ºC. Expression of the 18S ribosomal gene was used as a reference gene to normalize expression. The variation of expression of each of the genes in the various groups was calculated in relation to the expression obtained in the WT mice following the following formula:
Expression of the gene of interest = 2- ΔΔCt, where ΔΔCt = ΔCt treatment - ΔCt baseline31,32. All determinations were made in duplicate.
Osteocyte count
The tibiae of the mice were also extracted at the time of sacrifice and preserved in 10% formalin for later histological processing. Once processed and stained with hematoxylin-eosin, the osteocytes embedded in the trabeculae were counted in 2 to 4 non-serial sections corresponding to the region immediately below the epiphyseal plate of each of the tibiae. The number of osteocytes was normalized by their corresponding area of bone tissue calculated using the Image J program (ImageJ 15.3a, National Institutes of Health, USA), and following the recommendations of the American Society for Bone and Mineral Research33 . The number of resulting osteocytes was calculated by taking the mean of each count per mouse, and was expressed as the mean ± standard error of the mean (SEM).
In vitro cell cultures and viability assay
MC3T3-E1 murine osteoblast cells were cultured in alpha MEM medium supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich) and 1% penicillin-streptomycin. The stable line of murine MLOY-4 osteocytes was maintained in culture plates previously collagenized and with alpha MEM medium with 2.5% calf serum (Sigma-Aldrich), 2.5% FBS and 1% penicillin-streptomycin. Both cell lines were incubated at 17ºC in a 5% CO2 atmosphere. For viability assays, both cell lines were incubated in 75% subconfluence in 6-well plates and were pre-treated or not for one hour with PTN (5.5 nM), and subsequently incubated in the presence of 50 µM etoposide (apoptotic agent) in 1% fetal bovine serum for 48 hours. At 48 hours, the cells were counted, including those in the supernatant of the wells and the trypsin from each well, and a cell count was made with Trypan Blue 0.4% in PBS in the Neubauer chamber, distinguishing the living cells from the dead. The percentage of dead cells from each of the experiments was calculated. Three experiments were carried out with each condition in triplicate and the result was expressed as the mean ± standard error of the mean (SEM).
Statistics
The results were expressed as mean ± standard error of the mean (SEM). The comparison between various groups was made using the non-parametric Kruskall-Wallis test with a posteriori U Mann-Whitney test, if appropriate. A p<0.05 was considered significant. Analyzes were carried out with the Graphpad InStat software (San Diego, California, USA).
RESULTS
Gene expression in bone tissue in the absence of PTN
In the first place, we wanted to verify how the expression of genes related to bone metabolism varied in the absence of PTN and in the presence of inflammatory conditions after the administration of LPS. Among the expression of all the genes analyzed, we found that OCN was highly overexpressed in PTNKo mice (Figure 1A). However, the expression of an early osteocyte differentiation marker such as podoplanin (E11) did not undergo significant changes (Figure 1B). In the same way, the expression of the modulators of osteoblast and osteoclastic activity OPG and RANKL did not undergo significant changes in the mouse PTNKo (Figures 1C, 1D); the Wnt pathway inhibitor, Dkk1 (Figure 1E); VEGFR2 (Figure 1F); and the levels of PTHrP (Figure 1G) and Cox43 (Figure 1H). On the other hand, when normal WT mice were treated with LPS, we found that the levels of OCN, Dkk-1 and VEGFR2 were significantly decreased (Figures 1A, 1D, 1F). With respect to the PTNKo mice treated with LPS, we found a significant increase in the expression of E11 (Figure 1B), PTHrP (Figure 1G) and Cox43 (Figure 1H) with respect to the WT mice treated with LPS.
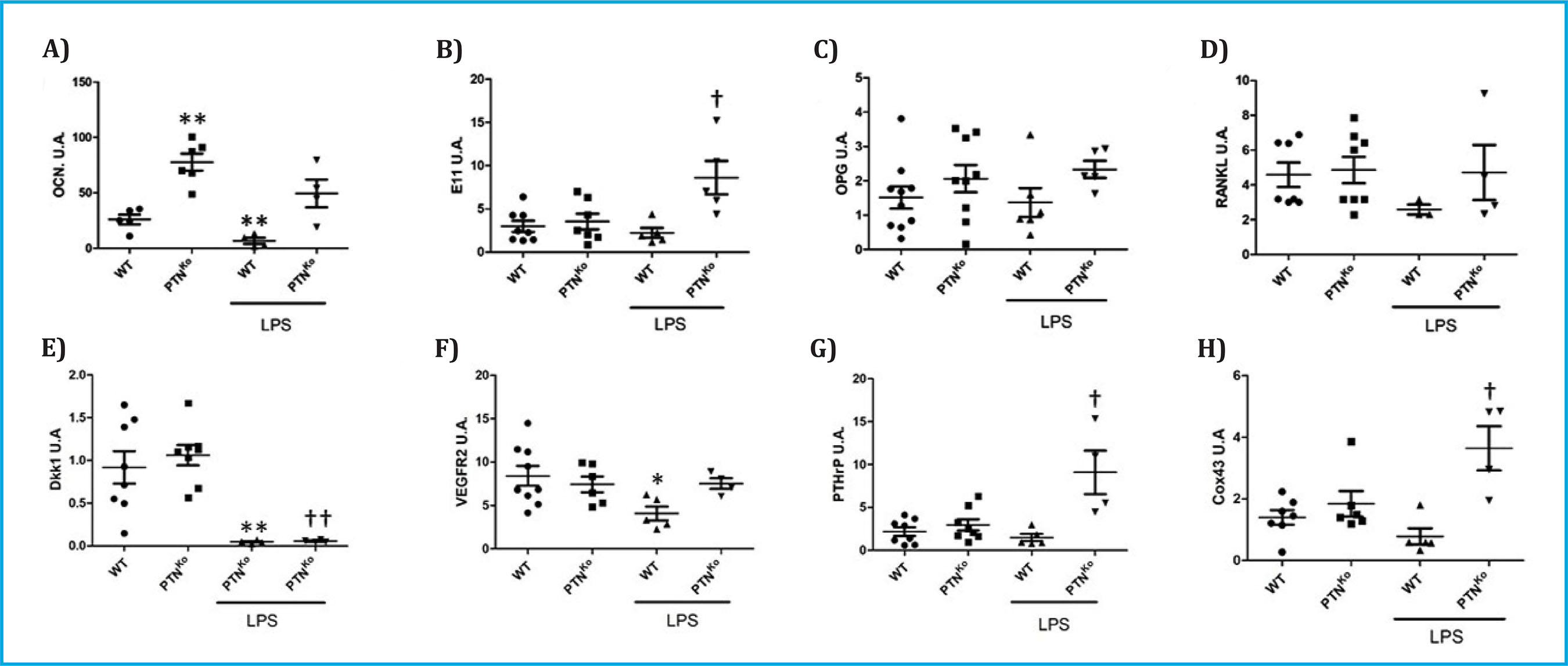
A) Osteocalcin (OCN) vs. WT, **p<0.05; B) Podoplanin (E11) vs. WT+LPS, †p<0.05; C) Osteoprotegerin (OPG); D) Activating receptor ligand for nuclear factor κB (RANKL); E) Dickoppf1 (Dkk1) vs. WT, **p<0.01; Dkk1 vs. WT+LPS, ††p<0.01; F) Vascular endothelial growth factor receptor 2 (VEGFR2) vs. WT, *p<0.05; G) Parathormone‐related peptide (PTHrP) vs. WT+LPS, †p<0.05; H) Conexin43 (Cox43) vs. WT+LPS, †p<0.05. The qPCR results are expressed in arbitrary units (A.U.) once normalized with the 18 S ribosomal control gene.
Figure 1. Expression of different genes in the vertebrae of WT or PTNKo mice treated or not with injection of LPS
The number of osteocytes was unchanged in PTNKo, WT+LPS and PTNKo+LPS mice
After observing that the only gene whose expression was regulated was OCN, a gene related to very advanced stages of osteoblastic maturation34. As this was upregulated, we thought that this expression could reflect an increase in the number of osteocytes in PTNKo mice. (We also extend this count to the WT and PTNKo groups treated with LPS). To do this, we stained the tibiae isolated from the mice and performed an exhaustive count of the osteocytes found in the tibial bone sections. As can be seen in Figures 2A, 2B, the number of osteocytes was similar in all groups of mice, not finding significant differences in any case.

A) Representative images of the tibia sections where the osteocyte count was carried out under the epiphyseal plate of the tibiae of the different experimental groups. B) Quantification of the number of osteocytes per mm2 of bone tissue in the WT, PTNKo, WT+LPS and PTNKo+LPS mice.
Figure 2. Number of osteocytes in the trabecular section of the tibiae of the WT and PTNKo mice
Protective effect of PTN against the induction of death in bone cells
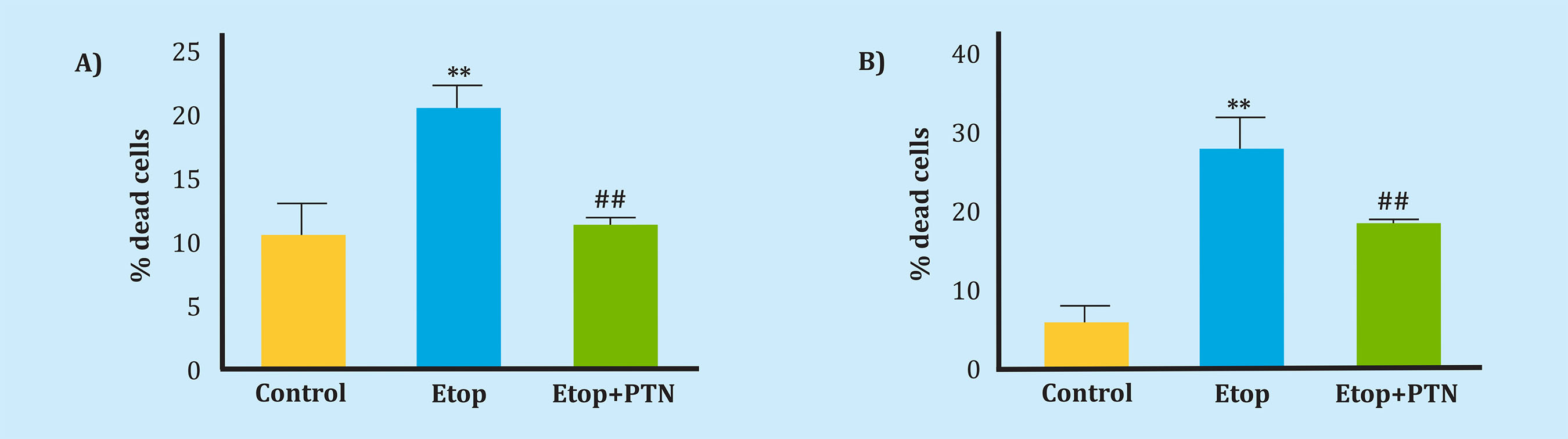
Given that the proliferative effect of PTN on MC3T3 osteoblasts has been previously described24, we wanted to verify the protective effect of PTN against the induction of death by etoposide in two murine cell lines, one of osteoblasts (MC3T3) and the other of osteocytes (MLOY-4), in vitro. The administration of PTN prior to treatment with etoposide produced a protective effect, since the cell death induced by this agent was significantly reduced in the MC3T3 line from 20.8% to 11.5% and in the MLOY4 line from 27, 5% to 18.1% (Figures 3A, 3B).
DISCUSSION
It is noteworthy that deleting a gene as important for the development and maintenance of bone tissue as PTN does not produce a phenotype marked by bone alterations. First of all, one might think that since PTN and Mdk have parallel expressions and similar functions, the lack of PTN could be compensated by the increase in Mdk. However, this does not appear to be the case, since it has been shown that Mdk-deficient mice have an increase in bone mass29, indicating that both cytokines do not share the same functions fully in bone tissue. In our study, we wanted to verify the expression of genes related to bone metabolism and whether this variation could explain, at least in part, the lack of bone effects in PTNKo mice.
Recent research indicates that OCN is a protein that acts by regulating the correct alignment of hydroxyapatite crystals with collagen, which is directly related to bone quality35, although it is also used as a marker of osteoblastic maturation36, and its current role as a boneproduced hormone is under debate37-39. The fact that it is increased in PTNKo mice led us to think that perhaps the number of osteocytes in parallel could be increased in these mice, as it is a marker of late maturation of the osteoblasts just before they become osteocytes. This may be one of the cellular mechanisms of compensation for the lack of PTN. However, our results (Figure 2A, 2B) show that the number of osteocytes does not vary significantly in the PTNKo mice compared to the WT mice, nor in the rest of the experimental groups. These results are consistent with those shown by Lehman et al, where an increase in OCN expression was observed in primary cultures from PTNKo mice and with an equivalent number of osteocytes between normal mice and PTNKo27.
The increase in osteocalcin probably has other effects on bone and the bone matrix of these mice presents an altered composition. To verify this, subsequent nuclear magnetic resonance studies would be necessary to analyze the quality and composition of the bone of the PTNKo mice. In contrast, and given the importance of PTN in certain inflammatory processes, we also wanted to investigate its role in the expression of these genes in the face of acute inflammation caused by injection of LPS. In this case, we discovered that the expression of E11, PTHrP and Cox43 was increased in the PTNKo mice compared to the WT mice treated with LPS. PTHrP is a cytokine that modulates bone remodeling, locally resembling the actions of parathormone40. This cytokine is involved in the inflammatory processes that occur in the bone marrow, promoting inflammatory factor MCP-1 expression in endothelial cells and osteoblasts41. E11 governs the first steps of the osteoblast-osteocyte transition42, its overexpression is a marker of bone tumors43 and of inflammation in other tissues and various tumors44. On the other hand, Cox43 is part of the communications established by osteocytes among themselves to form the osteocyte network. The role of connexins in inflammatory processes is diverse and variable depending on the different tissues45. Increases in the expression of Cox43 have been described that are related to diseases such as osteoarthritis46, and a greater expression in synoviocytes in response to stimuli such as LPS47. The decrease in Cox43 levels could be related to a decrease in arthritic inflammatory processes47. In our study, the absence of PTN, at least in the bone compartment, would have a detrimental role from an inflammatory point of view, since it would allow the overexpression of the three genes mentioned (Figures 1B, 1G, 1H) contrary to what occurs in the LPS-treated WT mouse, and thus would lead to undesired effects. Obviously, the ultimate verification of these observations requires subsequent studies both in vivo and in vitro to unravel the molecular mechanisms triggered by the absence of PTN in this context.
One of the limitations of this study is that the age of the mice (3 months), rather far from the 5 months in which a mature mouse can be considered from the skeletal point of view. It must be taken into account that murine models do not suffer from closure of the growth plates or estrogen depletion, which means that they always have little bone growth throughout their life. In this regard, it should be noted that severe bone defects have not been described in 50-week-old PTN-deficient mice27, although other authors have found small growth delays in 4-month-old mice28. In addition, in this latest study, it has been shown that the biomechanical characteristics of long bones are altered by having decreased the parameters of stiffness, resistance to breakage, and bone hardness28. Since PTN contributes to the correct alignment of the crystals of hydroxyapatite, future studies in the bones of adult or even aged mice with absence of PTN should include analysis of these bones by means of nuclear magnetic resonance to determine if the absence of PTN can cause brittle bone in advanced ages. Another study limitation is that, given that cortical areas have not been included when counting osteocytes, we cannot establish whether there are differences between the cortical and cancellous compartments, so this type of analysis must be carried out in subsequent studies.
Notably, it has been previously verified that PTN administration in vitro promotes the proliferation and differentiation of osteoblast cells24. Therefore, we wanted to verify, as far as we know for the first time, the protective effect of PTN against a pro-apoptotic stimulus such as etoposide in osteoblastic cells (MC3T3) and osteocytes (MLOY4), observing that PTN has a protective action. This result points to a possible additional role of PTN in situations of fracture or chronic inflammation in which it could protect, at least partially, bone cells. It is true that in this study the protective role that PTN can play in terms of death measured by LPS has not been proven. In this sense, previous studies have shown that this agent produces a senescent phenotype in osteocytes and an increase in bone resorption mediated by the production of IL-6 and RANKL by osteocytes, rather than death in a short period of time. as is the LPS administration time that has been carried out in this study. However, it would be interesting to study the role of PTN in these contexts in subsequent studies and in suitable models48,49.
Based on our findings, we can conclude that PTN deficiency is accompanied by an increase in OCN expression and that the induction of acute inflammation by means of LPS in PTN-deficient mice produces the overexpression of E11, PTHrP, and Cox43. Similarly, we have demonstrated the protective role of PTN in osteoblastic cells and osteocytes in the face of a cell death stimulus.
Funding: This research study was funded by the Ministry of Science, Innovation and Universities (RTI2018-095615-B-I00), the Community of Madrid (S2017/BMD-3684) and the FEIOMM grant for Translational Research 2015 "Impact of type 2 diabetes mellitus and the impact syndrome of type 2 diabetes mellitus and metabolic syndrome on the expression of miRNAs related to osteoarthritis”.
REFERENCES
1 Vanderwinden JM, Mailleux P, Schiffmann SN, Vanderhaeghen JJ. Cellular distribution of the new growth factor Pleiotrophin (HB-GAM) mRNA in developing and adult rat tissues. Anat Embryol (Berl). 1992;186:387-406. [ Links ]
2 Mitsiadis TA, Salmivirta M, Muramatsu T, Muramatsu H, Rauvala H, Lehtonen E, et al. Expression of the heparin-binding cytokines, midkine (MK) and HB-GAM (pleiotrophin) is associated with epithelial-mesenchymal interactions during fetal development and organogenesis. Development. 1995;121:37-51. [ Links ]
3 Li YS, Milner PG, Chauhan AK, Watson MA, Hoffman RM, Kodner CM, et al. Cloning and expression of a developmentally regulated protein that induces mitogenic and neurite outgrowth activity. Science. 1990;250:1690-4. [ Links ]
4 Merenmies J, Rauvala H. Molecular cloning of the 18-kDa growth-associated protein of developing brain. J Biol Chem. 1990;265:16721-4. [ Links ]
5 Tomomura M, Kadomatsu K, Matsubara S, Muramatsu T. A retinoic acidresponsive gene, MK, found in the teratocarcinoma system. Heterogeneity of the transcript and the nature of the translation product. J Biol Chem. 1990;265:10765-70. [ Links ]
6 Imai S, Kaksonen M, Raulo E, Kinnunen T, Fages C, Meng X, et al. Osteoblast recruitment and bone formation enhanced by cell matrix-associated heparin-binding growth-associated molecule (HB-GAM). J Cell Biol. 1998; 143:1113-28. [ Links ]
7 Rauvala H. An 18-kd heparin-binding protein of developing brain that is distinct from fibroblast growth factors. EMBO J. 1989;8:2933-41. [ Links ]
8 Tezuka K, Takeshita S, Hakeda Y, Kumegawa M, Kikuno R, Hashimoto-Gotoh T. Isolation of mouse and human cDNA clones encoding a protein expressed specifically in osteoblasts and brain tissues. Biochem Biophys Res Commun. 1990;173:246-51. [ Links ]
9 Kilpeläinen I, Kaksonen M, Kinnunen T, Avikainen H, Fath M, Linhardt RJ, et al. Heparin-binding growth-associated molecule contains two heparin-binding β-sheet domains that are homologous to the thrombospondin type I repeat. J Biol Chem. 2000;275: 13564-70. [ Links ]
10 Englund C, Birve A, Falileeva L, Grabbe C, Palmer RH. Miple1 and miple2 encode a family of MK/PTN homologues in Drosophila melanogaster. Dev Genes Evol. 2006;216:10-8. [ Links ]
11 Raulo E, Chernousov MA, Carey DJ, Nolo R, Rauvala H. Isolation of a neuronal cell surface receptor of heparin binding growth- associated molecule (HB-GAM). Identification as N-syndecan (syndecan-3). J Biol Chem. 1994; 269:12999-3004. [ Links ]
12 Deepa SS, Yamada S, Zako M, Goldberger O, Sugahara K. Chondroitin sulfate chains on syndecan-1 and syndecan-4 from normal murine mammary gland epithelial cells are structurally and functionally distinct and cooperate with heparan sulfate chains to bind growth factors: A novel function to control binding of midkine, pleiotrophin, and basic fibroblast growth factor. J Biol Chem. 2004;279:37368-76. [ Links ]
13 Mikelis C, Sfaelou E, Koutsioumpa M, Kieffer N, Papadimitriou E. Integrin α v β 3 is a pleiotrophin receptor required for pleiotrophin-induced endothelial cell migration through receptor protein tyrosine phosphatase β/ζ. FASEB J. 2009;23:1459-69. [ Links ]
14 Pantazaka E, Papadimitriou E. Chondroitin sulfate-cell membrane effectors as regulators of growth factor-mediated vascular and cancer cell migration. Biochim Biophys Acta. 2014; 1840:2643-50. [ Links ]
15 Said EA, Courty J, Svab J, Delbé J, Krust B, Hovanessian AG. Pleiotrophin inhibits HIV infection by binding the cell surface-expressed nucleolin. FEBS J 2005; 272:4646-4659. [ Links ]
16 Deuel TF. Anaplastic lymphoma kinase: ‘Ligand Independent Activation’ mediated by the PTN/RPTP?/? signaling pathway. Biochim Biophys Acta. 2013;1834:2219-23. [ Links ]
17 Schinke T, Gebauer M, Schilling AF, Lamprianou S, Priemel M, Mueldner C, et al. The protein tyrosine phosphatase Rptpzeta is expressed in differentiated osteoblasts and affects bone formation in mice. Bone. 2008;42:524-34. [ Links ]
18 Panicker N, Saminathan H, Jin H, Neal M, Harischandra DS, Gordon R, et al. Fyn kinase regulates microglial neuroinflammatory responses in cell culture and animal models of parkinson’s disease. J Neurosci 2015;35:10058-10077. [ Links ]
19 Xi G, Demambro VE, D’costa S, Xia SK, Cox ZC, Rosen CJ, et al. Estrogen stimulation of pleiotrophin enhances osteoblast differentiation and maintains bone mass in IGFBP-2 null mice. Endocrinology. 2020;161(4):bqz007. [ Links ]
20 Huang C, Ni Y, Wang T, Gao Y, Haudenschild CC, Zhan X. Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J Biol Chem. 1997;272:13911-5. [ Links ]
21 Perez-Pinera P, Berenson JR, Deuel TF. Pleiotrophin, a multifunctional angiogenic factor: Mechanisms and pathways in normal and pathological angiogenesis. Curr Opin Hematol. 2008; 15:210-4. [ Links ]
22 Rauvala H. An 18-kd heparin-binding protein of developing brain that is distinct from fibroblast growth factors. EMBO J 1989;8:2933-41. [ Links ]
23 Fernández-Calle R, Vicente-Rodríguez M, Gramage E, Pita J, Pérez-García C, FerrerAlcón M, et al. Pleiotrophin regulates microglia-mediated neuroinflammation. J Neuroinflammation. 2017;14(1):46. [ Links ]
24 Tare RS, Oreffo ROC, Clarke NMP, Roach HI. Pleiotrophin/osteoblast-stimulating factor 1: Dissecting its diverse functions in bone formation. J Bone Miner Res. 2002;17:2009-20. [ Links ]
25 Bouderlique T, Henault E, Lebouvier A, Frescaline G, Bierling P, Rouard H, et al. Pleiotrophin commits human bone marrow mesenchymal stromal cells towards hypertrophy during chondrogenesis. PLoS One. 2014;9:e88287. [ Links ]
26 Yang X, Tare RS, Partridge KA, Roach HI, Clarke NMP, Howdle SM, et al. Induction of human osteoprogenitor chemotaxis, proliferation, differentiation, and bone formation by osteoblast stimulating factor-1/pleiotrophin: osteoconductive biomimetic scaffolds for tissue engineering. J Bone Miner Res. 2003;18:47-57. [ Links ]
27 Lehmann W, Schinke T, Schilling AF, Catalá-Lehnen P, Gebauer M, Pogoda P, et al. Absence of mouse pleiotrophin does not affect bone formation in vivo. Bone. 2004;35:1247-55. [ Links ]
28 Imai S, Heino TJ, Hienola A, Kurata K, Büki K, Matsusue Y, et al. Osteocyte-derived HB-GAM (pleiotrophin) is associated with bone formation and mechanical loading. Bone. 2009;44:785-94. [ Links ]
29 Neunaber C, Catala-Lehnen P, Beil FT, Marshall RP, Kanbach V, Baranowsky A, et al. Increased trabecular bone formation in mice lacking the growth factor midkine. J Bone Miner Res. 2010; 25:1724-35. [ Links ]
30 Tavella S, Ruggiu A, Giuliani A, Brun F, Canciani B, Manescu A, et al. Bone turnover in wild type and pleiotrophin-transgenic mice housed for three months in the international space station (ISS). PLoS One. 2012;7:e33179. [ Links ]
31 Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-8. [ Links ]
32 Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101-8. [ Links ]
33 Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: A 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28:2-17. [ Links ]
34 Huang W, Yang S, Shao J, Li YP. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front Biosci. 2007;12:3068-92. [ Links ]
35 Nikel O, Laurencin D, McCallum SA, Gundberg CM, Vashishth D. NMR investigation of the role of osteocalcin and osteopontin at the organic-inorganic interface in bone. Langmuir. 2013;29:13873-82. [ Links ]
36 Weinreb M, Shinar D, Rodan GA. Different pattern of alkaline phosphatase, osteopontin, and osteocalcin expression in developing rat bone visualized by in situ hybridization. J Bone Miner Res. 1990;5:831-42. [ Links ]
37 Wei J, Karsenty G. An overview of the metabolic functions of osteocalcin. Rev Endoc. Metab Disord. 2015;16:93-8. [ Links ]
38 Diegel CR, Hann S, Ayturk UM, Hu JCW, Lim KE, Droscha CJ, et al. An osteocalcin-deficient mouse strain without endocrine abnormalities. PLoS Genet. 2020;16(5):e1008361. [ Links ]
39 Moriishi T, Ozasa R, Ishimoto T, Nakano T, Hasegawa T, Miyazaki T, et al. Osteocalcin is necessary for the alignment of apatite crystallites, but not glucose metabolism, testosterone synthesis, or muscle mass. PLoS Genet 2020;16 (5):e1008586. [ Links ]
40 Uy HL, Guise TA, Mata JD La, Taylor SD, Story BM, Dallas MR, et al. Effects of parathyroid hormone (pth)-related protein and pth on osteoclasts and osteoclast precursors in vivo. Endocrinology. 1995;136:3207-12. [ Links ]
41 Lu Y, Xiao G, Galson DL, Nishio Y, Mizokami A, Keller ET, et al. PTHrP-induced MCP-1 production by human bone marrow endothelial cells and osteoblasts promotes osteoclast differentiation and prostate cancer cell proliferation and invasion in vitro. Int J Cancer. 2007; 121:724-33. [ Links ]
42 Zhang K, Barragan-Adjemian C, Ye L, Kotha S, Dallas M, Lu Y, et al. E11/gp38 Selective Expression in Osteocytes: Regulation by Mechanical Strain and Role in Dendrite Elongation. Mol Cell Biol. 2006;26:4539-52. [ Links ]
43 Ariizumi T, Ogose A, Kawashima H, Hotta T, Li G, Xu Y, et al. Expression of podoplanin in human bone and bone tumors: New marker of osteogenic and chondrogenic bone tumors. Pathol Int. 2010;60:193-202. [ Links ]
44 Quintanilla M, Montero LM, Renart J, Villar EM. Podoplanin in inflammation and cancer. Int J Mol Sci. 2019;20 (3):707. [ Links ]
45 Willebrords J, Crespo Yanguas S, Maes M, Decrock E, Wang N, Leybaert L, et al. Connexins and their channels in inflammation. Crit Rev Biochem Mol Biol. 2016;51:413-39. [ Links ]
46 Casagrande D, Stains JP, Murthi AM. Identification of shoulder osteoarthritis biomarkers: Comparison between shoulders with and without osteoarthritis. J Shoulder Elb Surg. 2015; 24:382-90. [ Links ]
47 Tsuchida S, Arai Y, Kishida T, Takahashi KA, Honjo K, Terauchi R, et al. Silencing the expression of connexin 43 decreases inflammation and joint destruction in experimental arthritis. J Orthop Res. 2013;31:525-30. [ Links ]
48 Yu K, Ma Y, Li X, Wu X, Liu W, Li X, et al. Lipopolysaccharide increases IL-6 secretion via activation of the ERK1/2 signaling pathway to up-regulate RANKL gene expression in MLO-Y4 cells. Cell Biol Int. 2017;41:84-92. [ Links ]
49 Aquino-Martinez R, Rowsey JL, Fraser DG, Eckhardt BA, Khosla S, Farr JN, et al. LPS-induced premature osteocyte senescence: Implications in inflammatory alveolar bone loss and periodontal disease pathogenesis. Bone. 2020; 132:115-220. [ Links ]
Received: September 07, 2020; Accepted: November 24, 2020











 texto en
texto en