Meu SciELO
Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
International Microbiology
versão impressa ISSN 1139-6709
INT. MICROBIOL. vol.7 no.2 Jun. 2004
| RESEARCH ARTICLE | |||
|
| |||
| Peculiarities of the DNA of MM1, a temperate phage of Streptococcus pneumoniae
Summary. The abundant presence of temperate phages in the chromosomes of clinical isolates of Streptococcus pneumoniae has been well documented. The genome of MM1, a temperate phage of pneumococcus, has been isolated as a DNA-protein complex. The protein is covalently bound to the DNA, was iodinated in vitro with Na125I, and has an Mr of 22,000. Electron microscopy and enzymatic analyses revealed that the MM1 genome is a linear, circularly permuted, terminally redundant collection of double-stranded DNA molecules packaged via a headful mechanism. The location of the pac site appears to be downstream of the terminase, between orf32 and orf34 of the MM1 genome. [Int Microbiol 2004; 7(2):133-137]. [Int Microbiol 2004; 7(2):133-137] Key words: pneumococcus · DNA-protein complex · pac site · temperate phages | ||
| |||
Peculiaridades del DNA de MM1, un fago atemperado de Streptococcus pneumoniae Resumen. La abundante presencia de fagos atenuados en el cromosoma de aislados clínicos de Streptococcus pneumoniae ha sido bien documentada. El genoma de MM1, un fago atemperado de neumococo, se ha aislado como un complejo de DNA-proteína. La proteína unida covalentemente al DNA, fue yodada in vitro con Na125I, y tiene una Mr de 22.000. La microscopia electrónica y los análisis enzimáticos revelaron que el genoma de MM1 es una colección de moléculas lineales de DNA de doble cadena, permutadas circularmente y redundantes en la posición terminal, empaquetadas mediante un mecanismo de ensamblado de unidades genómicas discretas. También proponemos la localización del sitio pac en dirección 3´ de la terminasa, entre orf32 y orf34 del genoma de MM1. [Int Microbiol 2004; 7(2):133-137] Palabras clave: neumococo · complejo DNA-proteína · sitio pac · fagos atemperados | Peculiaridades do DNA de MM1, um fago atemperado de Streptococcus pneumoniae Resumo. Tem sido documentada uma abundante presença de fagos atenuados nos cromossomas de isolados clínicos de Streptococcus pneumoniae. O genoma de MM1, um fago atemperado de pneumococo, tem sido isolado como um complexo DNA-proteína. A proteína, unida covalentemente ao DNA, foi iodada com Na125I, e tem uma Mr de 22.000. A microscopia eletrônica e as análises enzimáticas revelaram que o genoma de MM1 é uma coleção de moléculas de DNA de dupla cadeia linear, permutadas circularmente, e redundantes na posição terminal, empacotadas mediante um mecanismo de ensamblado de unidades genômicas discretas. A localização do sitio pac parece estar na direção 3´ da terminasa, entre orf32 e orf34 do genoma de MM1. [Int Microbiol 2004; 7(2):133-137] Palavras chave: pneumococo · complexo DNA-proteína · sítio pac· fagos atemperados |
Introduction
Infectious diseases are currently the third leading cause of death in the USA, and Streptococcus pneumoniae is the most frequent cause of pneumonia and blood stream infections in the elderly, and one of the main causes of middle-ear infections in children. In some parts of the United States, resistance to penicillin in pneumococcus is as high as 20 to 40% [13]. Pneumococcal phages were first isolated in the mid-1970s [12,23]; since then, only 12 different phages have been identified and/or characterized [8,9]. Pneumococcal phages belong to four different morphological families and include lytic and temperate phages [8]. Lysogeny in pneumococci freshly isolated from humans was first documented by Bernheimer [2], who showed that about half of pneumococci recovered from pediatric patients and one third of isolates from adults yielded bacteriophages [1]. Moreover, recent surveys using the lytA gene as a probe have shown that many isolates contain prophages or some remnants of phage genomes [17]. Bernheimer [1] showed that 18 out of 20 multiply antibiotic-resistant pneumococcal strains of type 19A, first isolated in South Africa, were lysogenic whereas the other two strains lysed with mitomycin C, which suggested the presence of defective phages. She also showed that only one out of three antibiotic-sensitive 19A strains isolated from the same location was lysogenic. However, the indicator strain used in this assay was different from that employed for the drug-resistant strains. This leaves open the possibility of the presence of different phages in that analysis.
Several years ago, a phage named MM1 was isolated, and partially characterized [9], from the multi-resistant 23F Spanish pneumococcal clone (Spain23F-1) which is, together with South African strains, one of the best examples illustrating the rapid spread of drug resistance worldwide [15]. Microbial pathogens have developed a great variety of strategies to guarantee their own survival and expansion and, as already documented for many other bacterial species, phages might be important vehicles to introduce new factors that microorganisms can eventually use to cause infections and diseases [14]. Recently, a molecular analysis of MM1 was accomplished [16]. In this study, we report some peculiarities of the DNA of the first temperate phage of pneumococcus that has been completely sequenced.
Materials and methods
Bacteria, bacteriophage, and growth conditions. Streptococcus pneumoniae 949 strain, a member of the Spain23F-1 clone, was grown in C medium [11] supplemented with yeast extract (0.8 mg/ml; Difco laboratories) (C+Y medium) at 37ºC without shaking. Growth was monitored with a Hach 2100N nephelometer. Phage MM1 was induced from the lysogenic strain 949. At a cell concentration of 1.2 × 108 colony-forming units (CFU)/ml, mitomycin C was added to a final concentration of 75 ng/ml, and the culture was incubated in the dark at 37ºC until lysis occurred. The phages were precipitated with 0.5 M NaCl and 10% (w/v) polyethylene glycol 6000 and purified in a two-step CsCl gradient as previously described [6].
SDS-PAGE. SDS-polyacrylamide (10%) gel electrophoresis was done as described previously [22]. One part of the gel was stained with Coomassie blue and the other was dried for autoradiography.
DNA isolation. Pneumococcal DNA [5], proteinase K-treated MM1 DNA, and DNA-protein complexes [19] were prepared as previously described.
Electron microscopy. To visualize the DNA-associated protein by electron microscopy, a DNA-protein-complex solution (0.4 µg/ml) was treated with 0.1% glutaraldehyde in 10 mM magnesium acetate buffer, pH 7.0, according to a procedure published previously [21]. To test whether the MM1 genome had redundant ends, 2 µg proteinase K-treated phage DNA were denatured with 100 mM NaOH at room temperature for 15 min and renatured with formamide (35%, final concentration) as previously reported [21].
Radioactive labeling of DNA-protein complexes with 125I. DNA-protein complexes of MM1 were labeled with 125I to high specific activity by the chloramine-T method [10]. In short, to 20 µg DNA-protein complex in 100 µl SSC (0.15 M NaCl, 0.015 M sodium citrate), 20 µl TNET buffer [0.1 M Tris-HCl, pH 7.4, 2.5 M NaCl, 50 mM EDTA, 0.05% (v/v) Triton X-100] were added, followed by 7 µl Na125I (100 mCi/ml) and 5 µl chloramine-T solution (1 mg/ml). The mixture was incubated with gentle shaking for a minute at room temperature and the reaction terminated by the addition of 10 µl each of sodium metabisulfite (6 mg/ml) and KI (1.4 g/ml). Afterwards, 1% SDS (final concentration) was added, and the mixture was heated at 65ºC for 5 min and diluted with 100 µl TES buffer (10 mM Tris-HCl, pH 7.4, 5 mM EDTA, 0.1% SDS). The labeled complex was then chromatographed on a prepacked Sephadex G-25 column (Amersham-Pharmacia-Biotech, AP) equilibrated with TES buffer. The labeled fractions that eluted in the void volume were pooled, precipitated with ethanol and NaCl and dissolved in 10 mM Tris-HCl, pH 7.4, 1 mM EDTA.
Results and Discussion
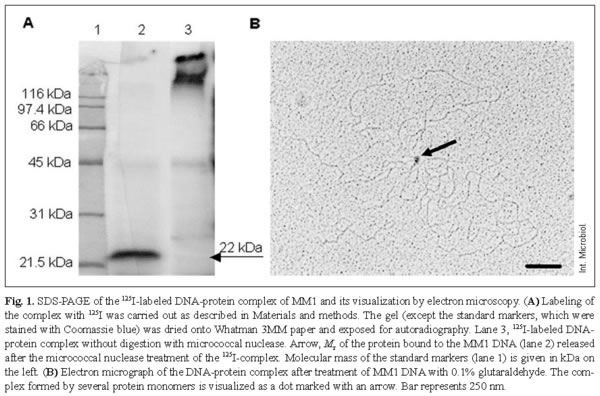
It was previously reported that MM1 DNA extracted from mature phage particles had been isolated as a DNA-protein complex [9]. To further study the characteristics of the protein associated with the DNA, the complex was labeled with 125I and purified as described in Materials and methods. The radioactivity was mainly found in the protein, as based on the following: (i) SDS-PAGE of the complex yielded 125I-labeling only at the top of the gel (Fig. 1A, lane 3), whereas no labeled band was found when proteinase K-treated complex was used (not shown); (ii) more than 95% of the 125I-radioactivity was solubilized by proteinase-K treatment, whereas almost 100% of the label remained acid-insoluble after incubation with either pancreatic DNAse or micrococcal nuclease. Electrophoresis of the 125I-labeled complex treated with micrococcal nuclease showed a major radioactive peak corresponding to an Mr of 22,000 (Fig. 1A, lane 2). Furthermore, when the complex was denatured by boiling in SDS, and β-mercaptoethanol, or urea, and analyzed by SDS-PAGE, no radioactivity migrated beyond the top of the gel (not shown), indicating that the 22-kDa protein is covalently linked to MM1 DNA. This protein was made visible by treating the complex with glutaraldehyde, which cross-links the protein monomers, and examined by electron microscopy. The monomeric molecules of the DNA-protein complex appear as a dot in Fig. 1B.
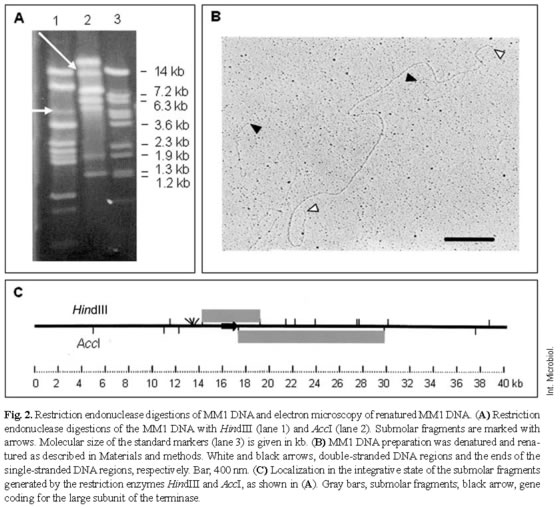
Restriction enzyme digestions followed by heating and rapid cooling failed to provide evidence for the presence of cohesive ends. Electron-microscopy measurements of a score of MM1 DNA molecules using pUC19 as an internal control provided an estimated average length of 42,240 ± 920 bp. As the MM1 prophage is 40,248-bp long [16], it was assumed that the MM1 phage particle packaged about 5 ± 2% more DNA than the genome unit, suggesting that MM1 has a redundant genome. Restriction endonuclease digestions of MM1 DNA with AccI or HindIII (Fig. 2A) revealed the presence of submolar fragments. These fragments may derive from the permuted terminal region of the MM1 DNA and would contain a specific site (pac) for initial recognition and cutting of phage concatemers sequentially packaged by the terminase enzyme [3]. It was also found that, for these restriction enzymes, the sizes of the submolar fragments agreed with those of the fragments predicted by computer analysis of the sequence. It is worth mentioning that submolar fragments are due to the peculiar headful packaging mechanism of this phage that generates the redundant genome, i.e. once the first pac site of the concatemer initiates the packaging process, the following encapsulated genome is excised downstream of the concatemer to the right of the subsequent pac site. These findings suggest that the pac site is located downstream of the gene encoding the large subunit of the terminase, between orf32 and orf34 [16]. Phage genes involved in DNA packaging are often located near the site for packaging, e.g. in phage P22 the pac site has been located within gene 3, coding for one element of the terminase [24]. These features also suggested that the MM1 genome is a terminally redundant and circularly permuted DNA generated by a headful packaging mechanism. To further address this point, MM1 DNA was denatured, renatured, and the re-associated molecules were analyzed by electron microscopy. Due to genome redundancy and permutation, all phage DNA molecules are not identical at their ends and, therefore, when non-identical genome strands are re-associated, chimeric DNA double-stranded molecules showing single-stranded ends with a variable size equivalent to the permutation length should be generated. As shown in Fig. 2B, the ends of the renatured molecules were single-stranded, as expected for a genome that is circularly permuted. Measurement of 20 complete DNA molecules revealed a circular permutation of about 50% of a genome unit, assuming that the concatemer formed during replication was built by 10 DNA units [3].
These results demonstrate that the phage genome contains a protein firmly bound to the DNA of the mature particle. Electrophoresis of the 125I-labeled complex treated with micrococcal nuclease revealed a major peak of radioactivity corresponding to an Mr of 22,000. It has been proposed that terminal proteins (TPs) from several well studied viruses serve as primers for the initiation of linear DNA replication [7,18,21]. However, it has been reported that DNAs of adenoviruses, and S-1 and S-2 plasmids from maize pollen cells, which also contain TPs, are capable of integrating into and excising from chromosomal or mitochondrial DNA-like mobile genetic elements [20]. Furthermore, Agrobacterium tumefaciens offers an example in which free, linear double-stranded T-DNA molecules generated from the Ti plasmid become covalently linked to a protein (VirD2) before being transferred to the genome of the host plant [4]. Taking into account that MM1 has a redundant genome with variable ends and most probably replicates by the rolling circle mechanism, leading to the formation of concatemers, a role of the 22-kDa protein covalently linked to MM1 DNA in replication is unlikely. Repeated attempts to determine whether the TP is bound to one or both DNA ends have been unsuccessful. Concerning the physiological role of such TPs, it is tempting to speculate that they link to the monomeric mature DNA once the terminase has cut down the concatemer and, thus, the TP might be involved in the packaging of the virion. However, other roles for TPs, such as a protective effect on DNA ends just after the infection or a putative influence on the DNA circularization (length reduction) step that should precede phage integration, cannot be ruled out. Note that genome redundancy is a critical factor to guarantee the integration of a complete copy of the phage genome. Redundancy facilitates a precise circularization by homologous recombination that is independent of the packaged-DNA length and of the nature of the terminal sequences. However, TP has to be removed during the circularization step to avoid any interference with the integration process.
In summary, because of the relevance of S. pneumoniae as a human pathogen and due to the abundant presence of temperate phages in the genomes of clinical isolates, a better knowledge of the molecular peculiarities of these phages might provide major tools to evaluate the involvement of pneumococcal phages in host virulence.
Acknowledgements. We acknowledge the help and advice of M. Rejas in electron microscopy. This work was supported by grants from the Dirección General de Investigación Científica y Técnica (BCM2000-1002) and from Programa de Grupos Estratégicos de la Comunidad Autónoma de Madrid.
References
1. Bernheimer HP (1979) Lysogenic pneumococci and their bacteriophages. J Bacteriol 138:618-624 [ Links ]
2. Bernheimer HP, Tiraby J-G (1976) Inhibition of phage infection by pneumococcus capsule. Virology 73:308-309 [ Links ]
3. Black LW (1989) DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol 43:267-292 [ Links ]
4. Durrenberger FA, Crameri A, Hohn B, Koukolikova-Nicola Z (1989) Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc Natl Acad Sci USA 86:9154-9158 [ Links ]
5. Fenoll A, Muñoz R, García E, de la Campa AG (1994) Molecular basis of the optochin-sensitive phenotype of pneumococcus: characterization of the genes encoding the F0 complex of the Streptococcus pneumoniae and Streptococcus oralis H+-ATPases. Mol Microbiol 12:587-598 [ Links ]
6. García E, Gómez A, Ronda C, Escarmís C, López R (1983) Pneumococcal bacteriophage Cp-1 contains a protein bound to the 5´ termini of its DNA. Virology 128:92-104 [ Links ]
7. García P, Hermoso JM, García E, García JL, López R (1986) Formation of a covalent complex between the terminal protein of pneumococcal bacteriophage Cp-1 and 5´-dAMP. J Virol 58:31-35 [ Links ]
8. García P, Martín AC, López R (1997) Bacteriophages of Streptococcus pneumoniae: a molecular approach. Microb Drug Resist 3:165-176 [ Links ]
9. Gindreau E, López R, García P (2000) MM1, a temperate bacteriophage of the type 23F Spanish/USA multiresistant epidemic clone of Streptococcus pneumoniae: structural analysis of the site-specific integration system. J Virol 74:7803-7813 [ Links ]
10. Greenwood FC, Hunter WM, Glover JS (1963) The preparation of 131I-labeled human growth hormone oh high specific radioactivity. Biochem J 89:114-123 [ Links ]
11. Lacks S, Hotchkiss RD (1960) A study of the genetic material determining an enzyme activity in Pneumococcus. Biochim Biophys Acta 39:508-517 [ Links ]
12. McDonnell M, Ronda-Laín C, Tomasz A (1975) Diplophage: a bacteriophage of Diplococcus pneumoniae. Virology 63:577-582 [ Links ]
13. McGee LK, Klugman KP, Tomasz A (2000) Serotypes and clones of antibiotic-resistant pneumococci. In A. Tomasz (ed) Streptococcus pneumoniae. Molecular biology and mechanism of disease. Mary Ann Liebert, Inc. Pub., Larchmont, New York, pp 375-379 [ Links ]
14. Miao EA, Miller SM (1999) Bacteriophages in the evolution of pathogen-host interaction. Proc Natl Acad Sci USA 96:9452-9454 [ Links ]
15. Muñoz R, Coffey TJ, Daniels M, Dowson CG, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser JM, Spratt BG, Tomasz A (1991) Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis 164:302-306 [ Links ]
16. Obregón V, García JL, García E, López R, García P (2003) Genome organization and molecular analysis of the temperate bacteriophage MM1 of Streptococcus pneumoniae. J Bacteriol 185: 2362-2368 [ Links ]
17. Ramirez M, Severina E, Tomasz A (1999) A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J Bacteriol 181:3618-3625 [ Links ]
18. Rekosh D, Russel WC, Bellet AJD, Robinson AJ (1977) Identification of a protein linked to the ends of adenovirus DNA. Cell 11:283-295 [ Links ]
19. Romero A, López R, Lurz R, García P (1990) Temperate bacteriophages of Streptococcus pneumoniae that contain protein covalently linked to the 5´ ends of their DNA. J Virol 64:5149-5155 [ Links ]
20. Salas M (1991) Protein-priming of DNA replication. Annu Rev Biochem 60:39-71 [ Links ]
21. Salas M, Mellado RP, Viñuela E, Sogo JM (1978) Characterization of a protein covalently linked to the 5´ termini of the DNA of Bacillus subtilis phage Φ29. J Mol Biol 119:269-291 [ Links ]
22. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [ Links ]
23. Tiraby JG, Tiraby E, Fox MS (1975) Pneumococcal bacteriophages. Virology 68:566-569 [ Links ]
24. Wu H, Sampson L, Parr R, Casjens S (2002) The DNA site utilized by bacteriophage P22 for initiation of DNA packaging. Mol Microbiol 45:1631-1646 [ Links ]