Meu SciELO
Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
International Microbiology
versão impressa ISSN 1139-6709
INT. MICROBIOL. vol.7 no.3 Set. 2004
| RESEARCH REVIEW | |||
|
| |||
| Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility
Summary. Although most organisms have detoxification abilities (i.e mineralization, transformation and/or immobilization of pollutants), microorganisms, particularly bacteria, play a crucial role in biogeochemical cycles and in sustainable development of the biosphere. Next to glucosyl residues, the benzene ring is the most widely distributed unit of chemical structure in nature, and many of the aromatic compounds are major environmental pollutants. Bacteria have developed strategies for obtaining energy from virtually every compound under oxic or anoxic conditions (using alternative final electron acceptors such as nitrate, sulfate, and ferric ions). Clusters of genes coding for the catabolism of aromatic compounds are usually found in mobile genetic elements, such as transposons and plasmids, which facilitate their horizontal gene transfer and, therefore, the rapid adaptation of microorganisms to new pollutants. A successful strategy for in situ bioremediation has been the combination, in a single bacterial strain or in a syntrophic bacterial consortium, of different degrading abilities with genetic traits that provide selective advantages in a given environment. The advent of high-throughput methods for DNA sequencing and analysis of gene expression (genomics) and function (proteomics), as well as advances in modelling microbial metabolism in silico, provide a global, rational approach to unravel the largely unexplored potentials of microorganisms in biotechnological processes thereby facilitating sustainable development. [Int Microbiol 2004; 7(3):173-180] Key words: aromatic compounds · catabolism · transcriptional regulation · metabolic engineering | ||
| |||
| Degradación bacteriana de contaminantes aromáticos: un paradigma de variabilidad metabólica Resumen. Aunque la mayoría de los organismos pueden detoxificar el ambiente (mediante procesos de mineralización, transformación y/o inmovilización de los contaminantes), los microorganismos, especialmente las bacterias, desempeñan un papel esencial en los ciclos biogeoquímicos y en el desarrollo sostenible de la biosfera. Después de los residuos glucosilados, el anillo de benceno es la unidad química estructural más frecuente en la naturaleza, y muchos compuestos aromáticos son contaminantes importantes. Las bacterias han desarrollado estrategias para obtener energía de todo tipo de compuestos mediante procesos aeróbicos o anaeróbicos (utilizando aceptores finales de electrones alternativos como los iones nitrato, sulfato y férrico). Los grupos de genes que intervienen en el catabolismo de compuestos aromáticos suelen localizarse en elementos genéticos móviles, tales como transposones y plásmidos. Dicha localización facilita su transferencia a otros organismos y, por tanto, la rápida adaptación de los microorganismos a nuevos contaminantes. Una estrategia que se ha empleado con éxito en procesos de biorremediación in situ es la combinación, en una única cepa bacteriana o en un consorcio microbiano, de diferentes capacidades degradadoras con otras características genéticas que aporten alguna ventaja selectiva en un ambiente determinado. Los métodos de alto rendimiento de secuenciación de DNA y de análisis global de la expresión génica (genómica) y funcional (proteómica), junto con los avances en los modelos in silico del metabolismo microbiano, proporcionan un enfoque global y racional para conocer las enormes posibilidades, aún inexploradas en su mayor parte, de la utilización de los microorganismos en procesos de biotecnología ambiental que faciliten el desarrollo sostenible. [Int Microbiol 2004; 7(3):173-180] Palabras clave: compuestos aromáticos · catabolismo · regulación transcripcional · ingeniería metabólica | Degradação bacteriana de contaminantes aromáticos: um paradigma de variabilidade metabólica Resumo. Muito embora a maioria dos organismos tenham a capacidade de detoxificar o ambiente (mediante processos de mineralización, transformação e/ou imobilização dos contaminantes), os microrganismos, especialmente as bactérias, desempenham um papel essencial nos ciclos biogeoquímicos e no desenvolvimento sustentável da biosfera. Depois dos resíduos glicosilados, o anel benzênico é a unidade química estrutural mais freqüente na natureza, e muitos compostos aromáticos são contaminantes importantes. As bactérias desenvolveram estratégias para obter energia, de todo tipo de compostos, mediante processos aeróbicos ou anaeróbicos (utilizando aceptores finais de elétrons alternativos como os ions nitrato, sulfato e férrico). Os grupos de genes que participam no catabolismo de compostos aromáticos estão localizados somente em elementos genéticos móveis, tais como os transposons e os plasmídeos. Esta localização facilita sua transferência para outros organismos e, portanto, a rápida adaptação aos novos contaminantes. Uma estratégia empregada com êxito nos processos de biorremediação in situ é a combinação, em uma única estirpe bacteriana ou em um consórcio microbiano, de diferentes capacidades de degradação com outras características genéticas que tragam alguma vantagem seletiva em um ambiente determinado. Os métodos de alto rendimento de sequenciamento de DNA e de análise global da expressão gênica (genômica) e funcional (proteomica), junto com os avanços nos modelos in silico do metabolismo microbiano, proporcionam um enfoque global e racional para conhecer as enormes possibilidades, ainda inexploradas em grande parte, da utilização dos microrganismos nos processos de biotecnologia ambiental que facilitem o desenvolvimento sustentável. [Int Microbiol 2004; 7(3):173-180] Palavras chave: Streptococcus pneumoniae · pneumococo · cápsula · bacteriófagos de pneumococo · enzimas líticas |
This article is based on the closing lecture of the 19th National Congress of the Spanish Society for Microbiology (SEM) given by the author in Santiago de Compostela, Spain, on September 25, 2003, on the occasion of receiving the 10th SEM Biennial Prize.
Introduction
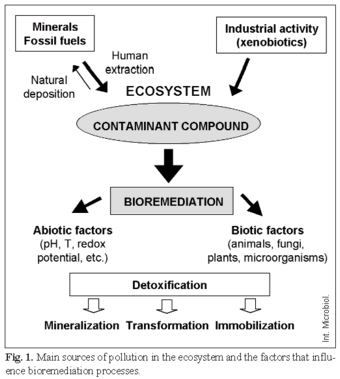
Environmental pollutants are compounds that are toxic to living organisms; they are released into the ecosystem at high concentrations, usually as a consequence of human activities. Contaminants are either compounds of industrial origin that present chemical structures alien to the biosphere (xenobiotics), e.g. polychlorobiphenyls (PCBs), polichlorodioxins, trinitrotoluene (TNT) and azo dyes, or natural compounds that have been mobilized to a bioavailable form that is toxic to organisms, e.g. hydrocarbons present in fossil fuels and heavy metals present in minerals (Fig. 1). Major sources of pollution are: (i) chemical and pharmaceutical industries that produce a wide array of xenobiotics and synthetic polymers; (ii) pulp and paper bleaching, which are the main sources of chlorinated organic compounds in the environment; (iii) mining, which releases heavy metals into biogeochemical cycles; (iv) fossil fuels (coal and petroleum), which may be accidentally released in large amounts into the ecosystem (oil spills) and whose combustion increases significantly CO2 atmospheric levels (green-house effect) and causes deposition of nitric and sulfuric acids (acid rain and smog); and (v) intensive agriculture, which releases massive amounts of fertilizers, pesticides, and herbicides [8,24].
The removal of pollutants from the environment via natural physico-chemical and biological processes (natural attenuation) is, in general, a slow and unpredictable way of counteracting anthropogenic pollution and irreversible damage to the biosphere. Therefore, the main, if not the only, successful strategy to fight pollution is the use and manipulation of the detoxification abilities of living organisms (bioremediation) (Fig. 1) [8,16,31,33,34]. Although most organisms are endowed with detoxification abilities, i.e mineralization, transformation and/or immobilization of pollutants, microorganisms, particularly bacteria, have been the most well-studied and the most frequently used for bioremediation strategies.
Bacteria, which evolved more than three billion years ago, have developed strategies for obtaining energy from virtually every compound. They play a crucial role in sustainable development of the biosphere and in biogeochemical cycles. The abundance of microorganisms, together with their great ability for horizontal gene transfer and their high growth rates, allows them to evolve quickly and to adapt to environmentally changing conditions, even to extreme environments that do not allow proliferation of other living organisms. The great genetic diversity of microorganisms accounts for their great metabolic versatility [6,16,27].
Microbial utilization of aromatic compounds
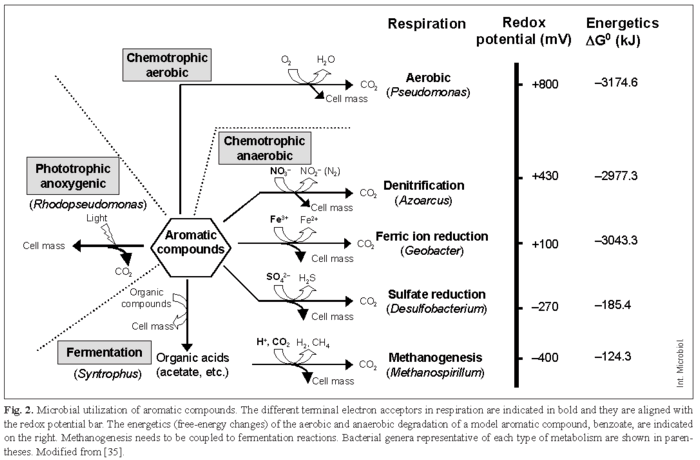
Next to glucosyl residues, the benzene ring is the unit of chemical structure most widely spread in nature. Moreover, the thermodynamic stability of the benzene ring increases its persistence in the environment; therefore, many aromatic compounds are major environmental pollutants [4]. By expressing different catabolic (biodegradative) pathways, microorganisms can use a wide array of aromatic compounds as sole carbon and energy sources [14]. The general ability of bacteria to use such compounds is related to the fact that most of these compounds are commonly present in the environment as a result of the recycling of plant-derived material [15]. Human-made xenobiotic compounds, by contrast, have been in contact with the microbiota only for about 100 years; therefore, some of them are still poorly degraded, if at all. Oxygen is the most common final electron acceptor for microbial respiration, and aerobic processes provide the highest amount of energy to cells (Fig. 2) [10]. In chemotrophic reactions, a portion of the substrate is oxidized to obtain energy and another part is assimilated into cell mass. In aerobic respiration, oxygen not only is the electron acceptor but also participates in activation of the substrate via oxygenation reactions.
Although a wide phylogenetic diversity of microorganisms is capable of aerobic degradation of contaminants, Pseudomonas species and closely related organisms have been the most extensively studied owing to their ability to degrade so many different contaminants [32]. However, many polluted environments are often anoxic, e.g., aquifers, aquatic sediments and submerged soils. In such environments, biodegradation is carried out by either strict anaerobes or facultative microorganisms using alternative electron acceptors, such as nitrate (denitrifying organisms), sulfate (sulfate reducers), Fe(III) (ferric-ion reducers), CO2 (methanogens), or other acceptors (chlorate, Mn, Cr, U, etc.) (Fig. 2) [13,16,35].
The use of electron acceptors other than oxygen is based on: (i) the electron-acceptor availability and (ii) the competition of different respiratory types of microorganisms for electron donors. For example, reduction of Fe(III) is the most frequent mechanism for oxidation of organic matter in subsurface environments. Sulfate is a major electron acceptor for the anaerobic degradation of contaminants in marine environments due to the high concentrations of sulfate in seawater [16]. In terms of energy, whereas degradation of aromatics using nitrate and Fe(III) as terminal electron acceptors is almost as efficient as that using oxygen, sulfate reducers and methanogenic conditions generate comparatively much less energy (Fig. 2) [10]. Consequently, the molar cell yields under methanogenic and sulfidogenic conditions are rather low. Fermentative strains tend to be restricted to a syntrophic existence because complete biodegradation becomes energetically favorable only when accompanying methanogens or sulfate-reducing bacteria promptly use the metabolic end-products generated during aromatic compound breakdown by fermenters (Fig. 2) [13]. Photosynthetic bacteria obtain energy from light and they degrade aromatic compounds anaerobically to form intermediary metabolites, such as acetyl-CoA, which are subsequently used in biosynthetic reactions (Fig. 2) [13]. Some aromatic compounds serve as electron acceptors rather than electron donors in bioremediation reactions. Thus, an important type of bioremediation is reductive dechlorination, in which bacteria remove chlorines from contaminants, such as chlorinated solvents and PCBs, by using these compounds as electron acceptors in dehalorespiration [9,13].
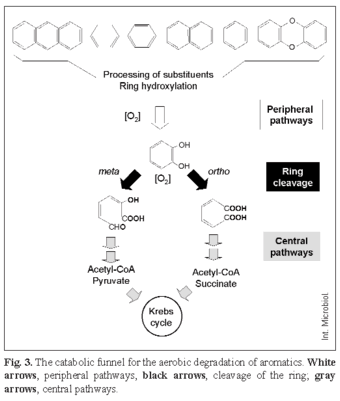
Aerobic and anaerobic biodegradation of aromatic compounds have several common features. Structurally diverse compounds are degraded through many different peripheral pathways to a few intermediates that are further channeled via a few central pathways to the central metabolism of the cell. In the aerobic catabolic funnel, most peripheral pathways involve oxygenation reactions carried out by monooxygenases and/or hydroxylating dioxygenases that generate dihydroxy aromatic compounds (catechol, protocatechuate, gentisate, homoprotocatechuate, homogentisate, hydroquinone, hydroxyquinol). These intermediate compounds are the substrates of ring-cleavage enzymes that use molecular oxygen to open the aromatic ring between the two hydroxyl groups (ortho cleavage, catalyzed by intradiol dioxygenases) or proximal to one of the two hydroxyl groups (meta cleavage, catalyzed by extradiol dioxygenases) (Fig. 3) [14].
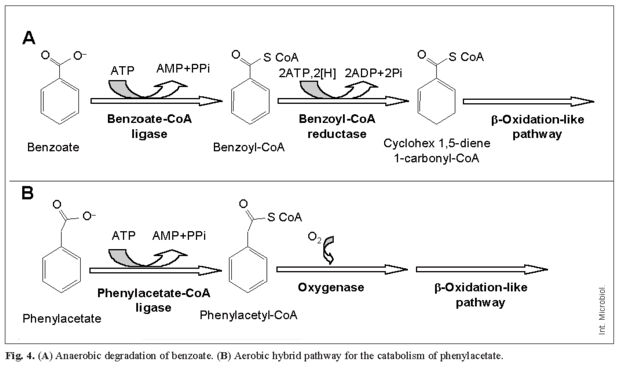
Central pathways involve a series of reactions leading to the formation of Krebs cycle intermediates (central metabolism). In the anaerobic catabolism of aromatic compounds, the peripheral pathways converge to benzoyl-CoA (occasionally to resorcinol and phloroglucinol), which becomes dearomatized by a specific multicomponent reductase that requires energy in the form of ATP (Fig. 4A) [13]. Finally, aerobic hybrid pathways, such as that for phenylacetic acid degradation, combine anaerobic features (initial formation of CoA derivatives) with typical aerobic reactions such as aromatic-ring oxygenation (Fig. 4B) [17]. Whereas pathways involved in degradation of xenobiotic compounds, such as those carrying halogen atoms or nitro groups, are only present in certain strains, those involved in the catabolism of common aromatic compounds, such as benzoate or aromatic amino acids, are widespread [23].
Genetics and evolution of biodegradation pathways
The genes responsible for biodegradation pathways are usually arranged in clusters that comprise: (i) catabolic genes encoding the enzymatic steps of the catabolic pathway; (ii) transport genes responsible for active uptake of the compound; and (iii) regulatory genes that adjust expression of the catabolic and transport genes to the presence of the compound to be degraded [7]. Bacteria thriving in the environment are exposed to a range of physical and chemical signals that need to be processed to achieve a positive or negative physiological response. For instance, bacteria are usually confronted with alternative carbon sources and they must "decide" which of them will be preferentially consumed before metabolizing less preferred substrates, such as pollutants, to guarantee, therefore, a satisfactory metabolic return. In order to achieve this goal, bacteria have developed a physiological response (superimposed regulation) that controls and adjusts the specific regulation of catabolic operons to the physiological and metabolic state of the cells [2]. The catabolic clusters are usually present in mobile genetic elements, such as transposons and plasmids, which facilitate their horizontal transfer of the respective genes and, therefore, rapid adaptation of microorganisms to the presence of new pollutants in a particular ecosystem [26,30].
Although microorganisms have acquired the ability to use pollutants as carbon and energy sources, their efficiency at removing such pollutants might not be optimal for cleaning up present-day pollution. In fact, microorganisms have evolved towards ecological fitness rather than biotechnological efficiency; thus, it would take a long time for bacteria capable of cleaning up anthropogenic pollution to evolve by natural selection. Hence, studying the physiology, biochemistry and genetics of the catabolic pathways becomes crucial to recreate and accelerate natural processes in the test tube as well as to accomplish their rational manipulation to design more efficient biocatalysts for different biotechnological applications. These include: (i) bioremediation of polluted sites, (ii) biotransformation of toxic compounds into fine chemicals and other high added-value products (green chemistry), and (iii) development of in situ biomonitoring devices and biosensors to monitor pollutant bioavailability [6,25,27]. The main limitations for the use of microorganisms in cleaning-up pollutants are summarized in Fig. 5, and some strategies to overcome these restrictions are presented below.
Designing recombinant bacteria for bioremediation
To enhance the metabolic efficiency of a microorganism for a particular environmental application, engineering should be carried out at two different levels: (i) manipulation of the specific catabolic pathway, and (ii) manipulation of the host cell. In order to improve the rate of pollutant removal and broaden the range of substrates of a catabolic pathway, manipulation of the key enzymes of both the pathway and the regulatory mechanisms that control the expression of the catabolic genes will be required [6,27]. Protein stability and protein activity can be altered and/or improved by protein engineering and rationally directed molecular evolution techniques (DNA shuffling and other in vitro recombination methods) [11,27]. Moreover, metabolic engineering allows the generation of novel hybrid pathways by assembling catabolic modules from different origins in the same host cell, thus leading to pathway expansion to new substrates, completion of incomplete pathways, the creation of new routes, and construction of bacteria with multiple pathways [20,23,24,27]. The rational combination of catabolic pathways may allow the complete metabolism of xenobiotics, as has been shown with the development of bacteria capable of mineralizing PCBs, and can prevent the formation of dead-end products and toxic metabolites by misrouting of the pollutants [20,27].
Bacteria used to remediate pollutants probably undergo environmental stress due to high concentrations of toxic contaminants, toxic solvents, extreme pH, temperature, ionic strength, etc. [27]. The combination in a single bacterial strain of different degradative abilities with genetic traits that provide selective advantages in the target site is a successful strategy for in situ bioremediation [20]. For instance, solvent-resistant strains may be ideal hosts to construct genetically engineered microorganisms for the removal of wastes with high solvent contents [20,22]. Heavy metals, including Hg, Cd and As, are currently major sources of pollution and cannot be destroyed or biodegraded. Recombinant microorganisms have been developed to accumulate and/or immobilize heavy metals present in soil and water [18,29]. As some metals can serve as terminal electron acceptors in microbial respiration, anaerobes that use them have been applied to reduce the soluble oxidized form of the metal to the insoluble form, thereby preventing its further spread in the environment [16]. Some polluted environments contain a mixture of organic wastes, heavy metals and high-energy radionuclides. In this context, radiation-resistant bacteria are being genetically engineered with biodegradation genes to render them suitable for the treatment of such mixed wastes [5,20,31].
One of the main reasons for the persistence of some aromatic compounds in the environment is their limited bioavailability. This is the case of hydrocarbons present in fossil fuels, which are frequently attached to soil particles. The host cell can be manipulated to enhance bioavailability of the pollutant by engineering the production of biosurfactants or by promoting chemotaxis of the biodegrader to the toxic compound [8,23]. Surfactant production has been combined with the ability to selectively cleave carbon-sulfur bonds in the sulfur-containing compounds present in oil (biodesulfurization) and, hence, efficient recombinant biodesulfurizers have been obtained [12]. Some anaerobes release Fe(III) chelators, which solubilize Fe(III) from Fe(III) oxides, and electron-shuttling compounds, which accept electrons from the cell surface and then reduce Fe(III) oxides. It has been shown that enhancing the availability of some electron acceptors, such as the insoluble Fe(III) oxides, by adding suitable ligands can greatly stimulate anaerobic degradation of contaminants in subsurface environments [16].
Biosafety is a major issue when releasing recombinant microorganisms into any open environment. In order to address this concern, several genetic circuits have been developed to allow survival of the recombinant microorganism only when present in the polluted site and during the time required for removal of the pollutant (biological containment). To avoid dispersal of the recombinant trait from the recombinant bacteria to the native microbial population, different gene-containment circuits based on a toxin and its cognate antidote have also been developed. Such active containment systems reduce significantly the potential risks that release of recombinant bacteria might cause in the ecosystem [21,28].
Developing syntrophic bacterial consortia for bioremediation
It is not always possible to combine in one only organism all the required features to solve a given environmental problem, as is the case for the removal of complex mixtures in sewage plants. Under these circumstances, a suitable strategy is the generation of syntrophic bacterial consortia whose members are specialized in certain biodegradative steps or in the degradation of certain pollutants [24,27]. Plant-microbe interactions are also a feasible alternative to remove toxic compounds from polluted environments (rhizoremediation). Bioremediation strategies involving transgenic plants that express bacterial genes for the immobilization of heavy metals or TNT removal (phytoremediation) are currently cost-effective green technologies of great public and scientific interest [20,34].
Ideally, bioremediation strategies would be designed based on knowledge of the microorganisms present in the polluted environments, their metabolic abilities, and how they respond to changes in environmental conditions. The advent of high-throughput methods for DNA sequencing and analysis of gene expression (whole-genome DNA microarrays) and function (proteomic techniques), as well as advances in the modeling of microbial metabolism (in silico biology), are providing new insights into environmental microbiology [16]. Biodegradation network models have been created that provide the basis for both predicting the abilities of existing or not-yet synthesized chemicals to undergo biodegradation and quantifying the evolutionary rate for their elimination in the future [19]. However, in practice, much of the required information is not yet available, and the use of microorganisms in bioremediation is highly empirical rather than knowledge-based. An alternative approach that has been shown to be successful for the treatment of some environmental disasters, including marine oil spills, is the addition of limiting nutrients (nitrogen, phosphorous, etc.) to the polluted site to enhance growth of the native microbiota able to degrade hydrocarbons (biostimulation) [34]. The addition of terminal electron acceptors, such as sulfates, has been shown to accelerate contaminant degradation by sulfate-reducer bacteria in aquifers [1].
Conclusion
The great versatility of microorganisms offers a simpler, inexpensive and more environmentally friendly strategy to reduce environmental pollution than non-biological options. Although microorganisms represent half of the biomass of our planet, we know as little as 5% of the microbial diversity of the biosphere [3], which reveals the largely unexplored genetic pool of the microbial world. Hence, the huge potential of environmental microbiology will become crucial for promoting the welfare and sustainable growth of our society.
Acknowledgements. Research in our laboratory is supported by grants 07M/0127/2000 and 07M/0076/2002 from the Comunidad Autónoma de Madrid, grants BIO2000-1076, BIO2003-01482, VEM2003-20075-CO2-02, and GEN2001-4698-CO5-02 from the CICYT, and by the EU contract QLRT-2001-02884.
References
1. Anderson RT, Lovley DR (2000) Anaerobic bioremediation of benzene under sulphate-reducing conditions in a petroleum-contaminated aquifer. Environ Sci Technol 34:2261-2266 [ Links ]
2. Cases I, de Lorenzo V (2001) The black cat/white cat principle of signal integration in bacterial promoters. EMBO J 20:1-11 [ Links ]
3. Curtis TP, Sloan WT, Scannell JW (2002) Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA 99:10494-10499 [ Links ]
4. Dagley S (1986) Biochemistry of aromatic hydrocarbon degradation in Pseudomonads. In: Sokatch J, Ornston LN (eds) The Bacteria. Academic Press, Orlando, Vol. 10, pp 527-555 [ Links ]
5. Daly MJ (2000) Engineering radiation-resistant bacteria for environmental biotechnology. Curr Opin Biotechnol 11:280-285 [ Links ]
6. De Lorenzo V (2001) Cleaning up behind us. EMBO Rep 2:357-359 [ Links ]
7. Díaz E, Prieto MA (2000) Bacterial promoters triggering biodegradation of aromatic pollutants. Curr Opin Biotech 11:467-475 [ Links ]
8. Dua M, Singh A, Sethunathan N, Johri AK (2002) Biotechnology and bioremediation: successes and limitations. Appl Microbiol Biot 59:143-152 [ Links ]
9. Fetzner S (1998) Bacterial dehalogenation. Appl Microbiol Biot 50:633-657 [ Links ]
10. Field JA, Stams AJM, Kato M, Schraa G (1995) Enhanced biodegradation of aromatic pollutants in cocultures of anaerobic and aerobic bacterial consortia. Antonie Leeuwenhoek 67:47-77 [ Links ]
11. Furukawa K (2000) Engineering dioxygenases for efficient degradation of environmental pollutants. Curr Opin Biotech 11:244-249 [ Links ]
12. Gallardo ME, Ferrández A, de Lorenzo V, García JL, Díaz E (1997) Designing recombinant Pseudomonas strains to enhance biodesulfurization. J Bacteriol 179:7156-7160 [ Links ]
13. Gibson J, Harwood CS (2002) Metabolic diversity in aromatic compound utilization by anaerobic microbes. Annu Rev Microbiol 56:345-369 [ Links ]
14. Harayama S, Timmis KN (1992) Aerobic biodegradation of aromatic hydrocarbons by bacteria. In: Sigel H, Sigel A (eds) Metal ions in biological systems. Marcel Dekker, New York, Vol. 28, pp 99-156 [ Links ]
15. Harwood CS, Parales RE (1996) The b-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol 50:553-590 [ Links ]
16. Lovley DR (2003) Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol 1:35-44 [ Links ]
17. Luengo JM, García JL, Olivera ER (2001) The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol Microbiol 39:1434-1442 [ Links ]
18. Mejáre M, Bülow L (2001) Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends Biotechnol 19:67-73 [ Links ]
19. Pazos F, Valencia A, de Lorenzo V (2003) The organization of the microbial biodegradation network from a systems-biology perspective. EMBO Rep 4:913 [ Links ]
20. Pieper DH, Reineke W (2000) Engineering bacteria for bioremediation. Curr Opin Biotech 11:262-270 [ Links ]
21. Ramos JL, Andersson P, Jensen LB, Ramos C, Ronchel MC, Díaz E, Timmis KN, Molin S (1995) Suicide microbes on the loose. Biotechnology 13:35-37 [ Links ]
22. Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-González MI, Rojas A, Teran W, Segura A (2002) Mechanisms of solvent tolerance in gram-negative bacteria. Annu Rev Microbiol 56:743-768 [ Links ]
23. Reineke W (1998) Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu Rev Microbiol 52:287-331 [ Links ]
24. Rieger PG, Meier HM, Gerle M, Vogt U, Groth T, Knackmuss HJ (2002) Xenobiotics in the environment: present and future strategies to obviate the problem of biological persistence. J Biotechnol 94:101-123 [ Links ]
25. Schmid A, Dordick JS, Hauer B, Kiener A, Wubbolts M, Witholt B (2001) Industrial biocatalysis today and tomorrow. Nature 409:258-268 [ Links ]
26. Tan H-M (1999) Bacterial catabolic transposons. Appl Microbiol Biotechnol 51:1-12 [ Links ]
27. Timmis KN, Pieper DH (1999) Bacteria designed for bioremediation. Trends Biotechnol 17:200-204 [ Links ]
28. Torres B, García JL, Díaz E (2004) Plasmids as tools for containment. In: Phillips G, Funnell B (eds) Plasmid Biology. ASM Press, Washington DC, pp 589-601 [ Links ]
29. Valls M, de Lorenzo V (2002) Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution. FEMS Microbiol Rev 26:327-338 [ Links ]
30. Van der Meer JR, de Vos WM, Harayama S, Zehnder AJB (1992) Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol Rev 56:677-694 [ Links ]
31. Wackett LP (2000) Environmental biotechnology. Trends Biotechnol 18:19-21 [ Links ]
32. Wackett LP (2003) Pseudomonas putida;—A versatile biocatalyst. Nat Biotechnol 21:136-138 [ Links ]
33. Wackett LP, Bruce NC (2000) Environmental biotechnology. Towards sustainability. Curr Opin Biotechnol 11:229-231 [ Links ]
34. Watanabe ME (2001) Can bioremediation bounce back? Nat Biotechnol 19:1111-1115 [ Links ]
35. Widdel F, Rabus R (2001) Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr Opin Biotechnol 12:259-276 [ Links ]