Meu SciELO
Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
International Microbiology
versão impressa ISSN 1139-6709
INT. MICROBIOL. vol.8 no.3 Set. 2005
| RESEARCH REVIEW | |||
|
| |||
| Biodegradation of lignocellulosics: microbial, chemical, and enzymatic aspects of the fungal attack of lignin
Summary. Wood is the main renewable material on Earth and is largely used as building material and in paper-pulp manufacturing. This review describes the composition of lignocellulosic materials, the different processes by which fungi are able to alter wood, including decay patterns caused by white, brown, and soft-rot fungi, and fungal staining of wood. The chemical, enzymatic, and molecular aspects of the fungal attack of lignin, which represents the key step in wood decay, are also discussed. Modern analytical techniques to investigate fungal degradation and modification of the lignin polymer are reviewed, as are the different oxidative enzymes (oxidoreductases) involved in lignin degradation. These include laccases, high redox potential ligninolytic peroxidases (lignin peroxidase, manganese peroxidase, and versatile peroxidase), and oxidases. Special emphasis is given to the reactions catalyzed, their synergistic action on lignin, and the structural bases for their unique catalytic properties. Broadening our knowledge of lignocellulose biodegradation processes should contribute to better control of wood-decaying fungi, as well as to the development of new biocatalysts of industrial interest based on these organisms and their enzymes. [Int Microbiol 2005; 8(3):195-204] Key words: wood-rotting fungi · lignin · analytical pyrolysis · oxidoreductases · catalytic mechanisms | ||
| |||
Biodegradación de la lignocelulosa: aspectos microbiológicos, químicos y enzimáticos del ataque fúngico a la lignina Resumen. La madera es el principal material renovable en la Tierra y es utilizada en gran parte como material de construcción y en la fabricación de celulosa. Esta revisión describe la composición de los materiales lignocelulósicos y diversos procesos de alteración de la madera por parte de hongos, como el deterioro causado por los llamados hongos de podredumbre blanca, de podredumbre parda y de podredumbre blanda y por los hongos cromógenos o manchadores de la madera. También se tratan los aspectos químicos, enzimáticos y moleculares del ataque a la lignina por los hongos, que es clave en el deterioro de la madera. Se describen las técnicas analíticas modernas para investigar la degradación y la modificación del polímero de la lignina causadas por hongos, así como las diversas enzimas oxidativas (oxidoreductasas) que intervienen en la degradación de la lignina. Entre éstas se encuentran lacasas, peroxidasas ligninolíticas de alto potencial redox (lignina-peroxidasa, manganeso-peroxidasa y peroxidasas versátiles) y las oxidasas. Se destacan las reacciones catalizadas, su acción sinérgica sobre la lignina, y las bases estructurales de sus exclusivas propiedades catalíticas. Un mejor conocimiento de los procesos de biodegradación de la lignocelulosa debería contribuir a un mejor control de los hongos descomponedores de la madera, así como al desarrollo de nuevos biocatalizadores de interés industrial basados en estos organismos y sus enzimas. [Int Microbiol 2005; 8(3):195-204] Palabras clave: hongos descomponedores de madera · lignina · pirólisis analítica · oxidoreductasas · mecanismos catalíticos | Biodegradação da lignocelulose: aspectos microbiológicos, químicos e enzimáticos do ataque fúngico à lignina Resumo. A madeira é o principal material renovável na Terra e é utilizada em grande parte como material de construção e na fabricação de celulose. Esta revisão descreve a composição dos materiais lignocelulósicos e diversos processos de alteração da madeira por parte de fungos, como a deterioração causada pelos chamados fungos de podridão branca, de podridão parda e de podridão mole e pelos fungos emboloradores ou manchadores da madeira. Também se tratam os aspectos químicos, enzimáticos e moleculares do ataque à lignina pelos fungos, que é chave na deterioração da madeira. Se descrevem as técnicas analíticas modernas para investigar a degradação e a modificação do polímero da lignina causadas por fungos, assim como as diversas enzimas oxidativas (oxidoreductasas) que intervêm na degradação da lignina. Entre estas se encontram lacasas, peroxidasas ligninolíticas de alto potencial redox (lignina-peroxidasa, manganeso-peroxidasa e peroxidasas versáteis) e as oxidasas. Se destacam as reações catalisadas, sua ação sinérgica sobre a lignina, e as bases estruturais de suas exclusivas propriedades catalíticas. Um melhor conhecimento dos processos de biodegradação da lignocelulosa deveria contribuir para um melhor controle dos fungos descomponedores da madeira, assim como ao desenvolvimento de novos biocatalisadores de interesse industrial baseados nestes organismos e suas enzimas. [Int Microbiol 2005; 8(3):195-204] Palavras chave: fungos descomponedores de madeira · lignina· pirólise analítica · oxidoreductasas · mecanismos catalíticos |
Lignocellulosic materials
Forests represent approximately 27% of the world's land area, and wood is the predominant commercial product from forests. Global wood consumption is around 3500 million m3/year, and has increased over 65% since 1960. More than half of this consumption is for fuel. The remainder of the global roundwood consumption is largely for pulp and paper products, building materials, and other wood in service.
Wood and other lignocellulosic materials are formed by three main polymeric constituents, cellulose, lignin, and hemicelluloses [25]. Cellulose is a linear and highly ordered (often crystalline) polymer of cellobiose (D-glucopyranosyl-β-1,4-D-glucopyranose) that represents over 50% of wood weight. By contrast, lignin is a three-dimensional network built up of dimethoxylated (syringyl, S), monomethoxylated (guaiacyl, G) and non-methoxylated (p-hydroxyphenyl, H) phenylpropanoid units, derived from the corresponding p-hydroxycinnamyl alcohols, which give rise to a variety of subunits including different ether and C-C bonds. Acetylated lignin units have been recently identified in non-woody plants using analytical pyrolysis [7].
Lignin is highly resistant towards chemical and biological degradation, and confers mechanical resistance to wood. The highest concentration of this recalcitrant polymer is found in the middle lamella, where it acts as a cement between wood fibers, but it is also present in the layers of the cell wall (especially the secondary cell-wall), forming, together with hemicelluloses, an amorphous matrix in which the cellulose fibrils are embedded and protected against biodegradation [14]. Lignin composition in terms of the H:G:S ratio varies between different vascular plant groups. Woody gymnosperms (softwoods) have the highest lignin content, and their lignin is made up mostly of G units. By contrast, lignin of woody angiosperms (hardwoods) consists of S and G units, and that from non-woody angiosperms contains also H units. Lignin composition between the different wood tissues and cell-wall layers also varies. For example, middle-lamella lignin typically has a lower S/G ratio than lignin from the secondary wall.
The third structural component, hemicelluloses (polyoses), has an intermediate degree of complexity and is made up of different pentose and hexose residues, which are often acetylated, and generally form branched chains. Typically, hemicelluloses in softwood are glucomannans, whereas those in hardwoods are mainly xylans together with variable percentages of galactose, arabinose, rhamnose and methylglucuronic acid units, and acetyl groups. Other non-structural components of wood include compounds extractable with organic solvents (the so-called extractives) which can be either polar (e.g. phenols and tannins) or apolar (e.g. fats and sterols), water-soluble compounds (e.g. sugars and starch), as well as proteins and ashes. These components together generally represent less than 5% of the dry weight of wood but can reach 20% in some softwoods (e.g. in some Cupressaceae)[14].
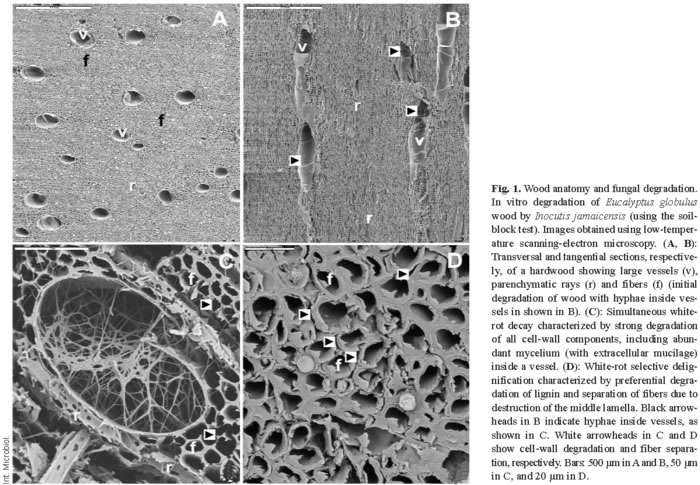
The above constituents form the three main types of wood tissue elements, namely fibers, vessels, and parenchyma cells (Fig. 1A,B). In gymnosperms, wood tissues have a relatively simpler structure than in angiosperms; they consist of 90-95% tracheid cells (softwood fibers) and low amounts of parenchyma, which includes the specialized resin channels in conifers. Parenchymatic rays, which contain phenolic and lipophilic extractives and water-soluble compounds as storage material, have a radial arrangement in wood. Vessels are large cells with a longitudinal arrangement, and they are responsible for the transport of water and nutrients along the plant stem. Finally, fibers, which are also longitudinally arranged, represent most of the wood volume and are characterized by their thick cell walls, which provide support to the tree.
Wood biodegradation
Lignocellulose degradation is a central step for carbon recycling in land ecosystems. Moreover, fungal decay of wood in service results in billion-euro losses. Basidiomycetes are the main wood rotters due to their ability to degrade or modify lignin, an enzymatic process that originated in the Upper Devonian period in parallel with the evolution of vascular plants [12]. Wood-rotting basidiomycetes are classified as white-rot and brown-rot fungi based mainly on macroscopic aspects [42,49]. Table 1 summarizes the characteristics of wood attack by several types of fungi. Basidiomycetes can overcome difficulties in wood decay, including the low nitrogen content of wood and the presence of toxic and antibiotic compounds. Extracellular oxidative enzymes (oxidoreductases) secreted by fungi are involved in degradation of cell-wall components (see below). White-rot basidiomycetes, the most frequent wood-rotting organisms, are characterized by their ability to degrade lignin, hemicelluloses, and cellulose, often giving rise to a cellulose-enriched white material. Due to the ability of white-rot basidiomycetes to degrade lignin selectively or simultaneously with cellulose, two white-rot patterns have been described in different types of wood, namely selective delignification, also called sequential decay, and simultaneous rot [35] (Fig. 1C,D). Brown-rot fungi, which grow mainly on softwoods, represent only 7% of wood-rotting basidiomycetes. This group of basidiomycetes can degrade wood polysaccharides after only a partial modification of lignin, resulting in a brown material consisting of oxidized lignin, which represents a potential source of aromatic compounds for the stable organic matter fraction in forest soils.
Although only white-rot and brown-rot basidiomycetes can degrade wood extensively, some ascomycetes and their asexual states, the so-called deuteromycetes, can colonize wood in contact with soil. This results in a decrease in the mechanical properties of wood, giving rise to so-called soft-rot, a process that often involves bacteria (Table 1). Soft-rot fungi can degrade wood under extreme environmental conditions (high or low water potential) that prohibit the activity of other fungi. Moreover, some basidiomycetes also cause a soft-rot-type decay pattern. Finally, a limited number of ascomycetous fungi, called stain fungi, can colonize wood through parenchymatic rays and resin channels causing discoloration of softwood tissues but a very limited degradation, which mainly affects extractives and water-soluble materials.
Chemical analysis of degraded wood
The main wood degradation patterns at advanced stages of decay can be identified macroscopically and microscopically, as described above. General changes in the chemical composition of wood can be also observed after fungal-induced decay (Table 1). However, a precise analysis of the degradation type requires chemical analysis of cellulose and lignin contents and of the modifications in the decayed wood. Lignin in wood is traditionally estimated by the Klason method, which is based on total acid hydrolysis of polysaccharides and gravimetric estimation of the lignin content (after deducing ashes and protein). This method, however, is time-consuming, and requires a considerable sample volume. In addition, its application to samples containing modified lignin (as in rotted wood samples) is problematic because it has been developed for lignin in sound (undecayed) wood. Estimation of Klason lignin is often combined with analysis of wood polysaccharide composition by gas chromatography (GC) of the monosaccharides present in the acid hydrolysate.
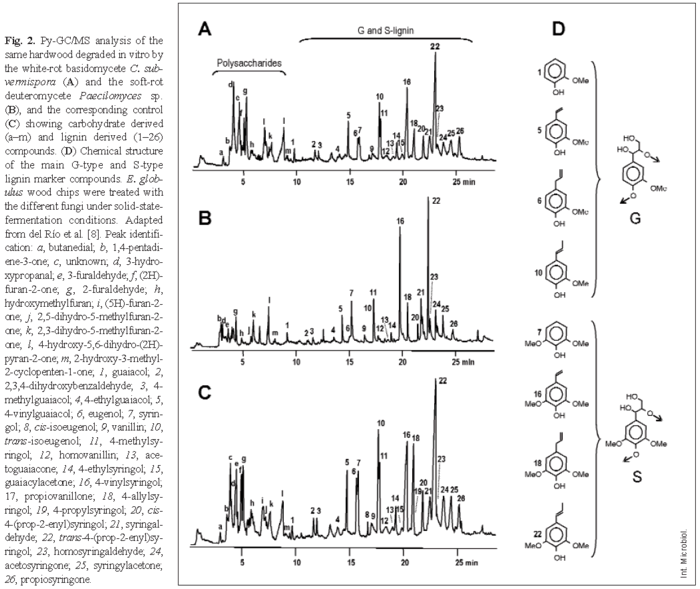
Several modern spectroscopic and degradative methods have been used to analyze lignin or polysaccharides in wood [30]. Among them, pyrolysis coupled to gas chromatography-mass spectrometry (Py-GC/MS) has several great advantages as a technique enabling rapid analysis of small samples, yielding a precise identification of the proportion of H, G, and S lignin units [8]. Figure 2 shows a Py-GC/MS analysis of the fungal decay of a hardwood (Eucalyptus globulus). Two regions, corresponding to the polysaccharide- and lignin-derived compounds, can be distinguished in the pyrograms. Due to the higher stability of the degradation products of lignin (the main ones shown in Fig. 2D) than of polysaccharides, which yield several compounds that cannot be chromatographied, Py-GC/MS analyses result in overestimation of lignin. However, this method can be used to compare the changes in the relative proportion of lignin and polysaccharides in decaying wood. Moreover, the amounts of G-lignin and S-lignin (no H-lignin has been detected in eucalypt wood) can be separately estimated by Py-GC/MS analysis.
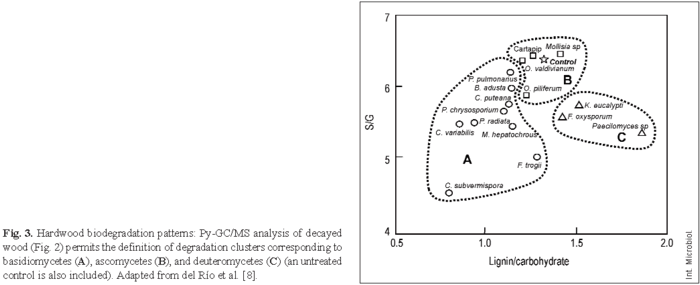
Figure 2A shows wood decay by the white-rot basidiomycete Ceriporiopsis subvermispora, which results in a relative decrease in the lignin peaks and a relative increase in the carbohydrate peaks. By contrast, the soft-rot deuteromycete Paecilomyces sp. decreases cellulose content to a larger extent than lignin content (Fig. 2B). Figure 3 shows wood degradation patterns by the basidiomycetes Bjerkandera adusta, C. subvermispora, Coniophora puteana, Crepidotus variabilis, Funalia trogii, Melanotus hepatochrous, Phanerochaete chrysosporium, Phlebia radiata, and Pleurotus pulmonarius (Fig. 3A), the ascomycetes Mollisia sp, Ophiostoma piliferum (including an inoculum commercialized as Cartapip) and Ophiostoma valdivianum (Fig. 3B), and the deuteromycetes Fusarium oxysporum, Kirramyces eucalypti and Paecilomyces sp. (Fig. 3C), which can be differentiated based on their lignin/carbohydrate and S/G Py-GC/MS ratios. The highest cellulose enrichments are produced by the basidiomycetes C. subvermispora and C. variabilis, the former also causing the highest modification of lignin, as revealed by the S/G ratio. In addition, Paecilomyces sp. and other deuteromycetes produce an increase in the relative lignin content in wood (due to preferential removal of polysaccharides), whereas ascomycetes slightly modify wood composition, as revealed by the Py-GC/MS analysis.
Enzymatic aspects
Lignin degradation and/or modification by basidiomycetes is the key step in lignocellulose decay. Therefore, the enzymes and mechanisms involved in lignin attack are described below. For a discussion of the subsequent steps in the degradation of wood polysaccharides, we recommend other reviews [36,43].
Laccases have been known for many years in plants, fungi, and insects, where they play a variety of roles, including synthesis of pigments, fruit-body morphogenesis, and detoxification [34]. Their production in fungal plate cultures was considered to be a characteristic unique to white-rot basidiomycetes [27], although some brown-rot fungi produce laccase in liquid cultures [29]. These phenoloxidases have a low redox potential that allows direct oxidation only of phenolic lignin units, which often comprise less than 10% of the total polymer. The interest in laccases for biotechnological applications increased with the discovery of their ability to oxidize high redox potential substrates in the presence of synthetic mediators [3], which allows the degradation of xenobiotic compounds [40] and chlorine-free bleaching of paper pulp [4]. Natural mediators involved in lignin biodegradation remain to be identified, although some lignin-derived phenols could act as efficient laccase mediators [5].
Lignin peroxidase (LiP) and manganese peroxidase (MnP) were discovered in the mid-1980s in P. chrysosporium and described as true ligninases because of their high redox potential [16,31]. LiP degrades non-phenolic lignin units (up to 90% of the polymer), whereas MnP generates Mn3+, which acts as a diffusible oxidizer on phenolic or non-phenolic lignin units via lipid peroxidation reactions [26]. More recently, versatile peroxidase (VP) has been described in Pleurotus [32,41] and other fungi as a third type of ligninolytic peroxidase that combines the catalytic properties of LiP, MnP, and plant/microbial peroxidases oxidizing phenolic compounds [24].
Other extracellular enzymes involved in wood lignin degradation are oxidases generating H2O2, and mycelium-associated dehydrogenases that reduce lignin-derived compounds. The former include the aryl-alcohol oxidase (AAO) described in Pleurotus eryngii [18] and other fungi, and P. chrysosporium glyoxal oxidase [28]. Fungal aryl-alcohol dehydrogenases (AAD) and quinone reductases (QR) are also involved in lignin degradation [20,21].
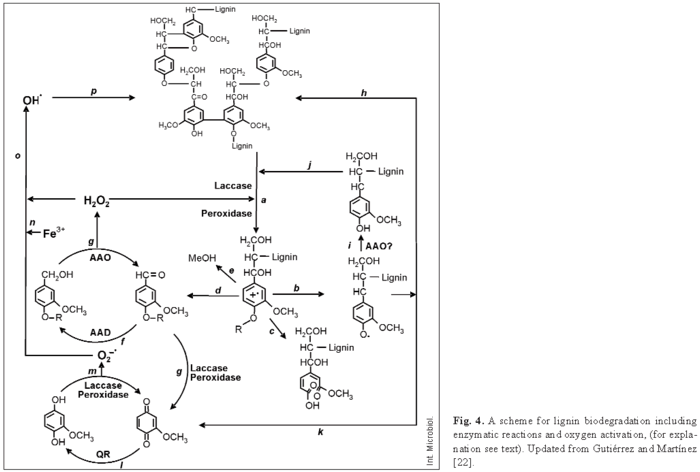
As shown in Fig. 4, laccases or ligninolytic peroxidases (LiP, MnP, and VP) produced by white-rot fungi oxidize the lignin polymer, thereby generating aromatic radicals (a) [12]. These evolve in different non-enzymatic reactions, including C4-ether breakdown (b), aromatic ring cleavage (c), Cα-Cβ breakdown (d), and demethoxylation (e) [22]. The aromatic aldehydes released from Cα-Cβ breakdown of lignin, or synthesized de novo by fungi (f, g) [21] are the substrate for H2O2 generation by AAO in cyclic redox reactions involving also AAD [19]. Phenoxy radicals from C4-ether breakdown (b) can repolymerize on the lignin polymer (h) if they are not first reduced by oxidases to phenolic compounds (i), as reported for AAO [33]. The phenolic compounds formed can be again reoxidized by laccases or peroxidases (j). Phenoxy radicals can also be subjected to Cα-Cβ breakdown (k), yielding p-quinones. Quinones from g and/or k contribute to oxygen activation in redox cycling reactions involving QR, laccases, and peroxidases (l, m) [20]. This results in reduction of the ferric iron present in wood (n), either by superoxide cation radical or directly by the semiquinone radicals, and its reoxidation with concomitant reduction of H2O2 to hydroxyl free radical (OH·) (o) [17]. The latter is a very strong oxidizer that can initiate the attack on lignin (p) in the initial stages of wood decay, when the small size of pores in the still-intact cell wall prevents the penetration of ligninolytic enzymes [13]. Then, lignin degradation proceeds by oxidative attack of the enzymes described above. In the final steps, simple products from lignin degradation enter the fungal hyphae and are incorporated into intracellular catabolic routes.
Molecular aspects
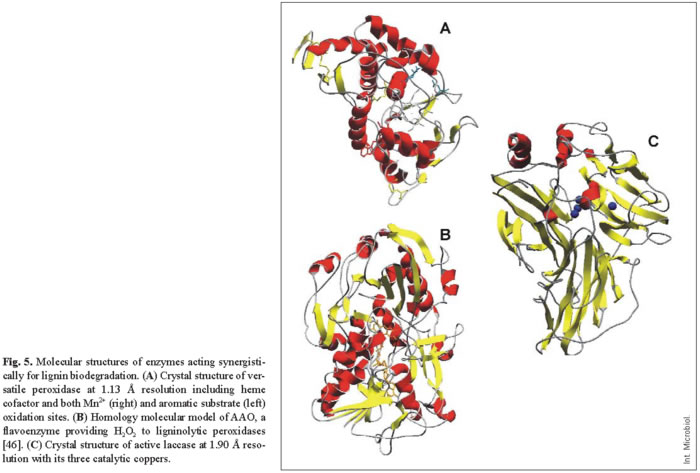
Due to their potential use as industrial biocatalysts, the catalytic mechanisms of lignin-degrading oxidoreductases (including peroxidases, oxidases, and laccases) have been extensively investigated and their molecular structures have been described (Fig. 5).
LiP and MnP were the second and third peroxidases whose crystal structure was solved [38,39,44], just 10 years after their discovery in P. chrysosporium. These peroxidases catalyze the oxidation of the recalcitrant non-phenolic lignin units by H2O2 . This is possible because of the formation of a high redox potential oxo-ferryl intermediate during the reaction of the heme cofactor with H2O2 . This two-electron reaction allows the activated enzyme to oxidize two substrate units, being reduced to the peroxidase resting state (which reacts again with peroxide). The catalytic cycle, consisting of the resting peroxidase and compounds I (two-electron oxidized form) and II (one-electron oxidized form), is common to other peroxidases. However, two aspects in their molecular structure provide ligninolytic peroxidases their unique catalytic properties: (i) a heme environment, conferring high redox potential to the oxo-ferryl complex; and (ii) the existence of specific binding sites (and mechanisms) for oxidation of their characteristic substrates, including non-phenolic aromatics in the cases of LiP, manganous iron in the case of MnP, and both types of compounds in the case of the new VP.
Similar heme environments in the above three peroxidases (located at the central region of the protein, Fig. 5A) have been evidenced by 1H-NMR, which allows the signals of both the heme cofactor protons and several amino acid residues forming the heme pocket to be identified [1]. This is possible due to the paramagnetic effect caused by the cofactor iron, which displaces the signals of neighbor protons outside the region where most protein protons overlap. One of the main differences observed among peroxidases is the position of a protein iron ligand, the Nε of the side-chain of a histidine residue (the so-called proximal histidine). In ligninolytic peroxidases, this residue is displaced away from the heme iron, increasing its electron deficiency and increasing the redox potential of the oxo-ferryl complex [31].
In addition, recent studies have contributed to identification of the substrate binding sites in ligninolytic peroxidases. The aromatic substrate binding site and the manganese binding site were first identified in LiP and MnP [9,16,45], and then confirmed in the crystal structure of VP solved at atomic resolution (unpublished). These studies revealed that the novel catalytic properties of VP are due to its hybrid molecular architecture, as suggested several years before [6,41]. Mn2+ oxidation is produced at a binding site near the cofactor, where this cation is bound by the carboxylates of three acidic residues, which enables direct electron transfer to one of the heme propionates (Fig. 5A right). By contrast, veratryl alcohol (and other lignin model substrates) are oxidized at the surface of the protein by a long-range electron transfer mechanism that initiates at an exposed tryptophan residue (Fig. 5A left). The rationale of the existence of this electron transfer mechanism is related to the fact that many LiP/VP aromatic substrates, including the lignin polymer, cannot penetrate inside the protein to transfer electrons directly to the cofactor. Therefore, these substrates are oxidized at the enzyme surface, and electrons are transferred to the heme by a protein pathway.
The H2O2 responsible for oxidative degradation of lignin is generated by extracellular fungal oxidases, which can reduce dioxygen to peroxide in a catalytic reaction. Flavin cofactors are generally involved in this reaction, as in the Pleurotus flavoenzyme AAO, although glyoxal oxidase from P. chrysosporium is a copper-containing oxidase [47]. Among flavoenzyme oxidases, fungal glucose oxidase has been crystallized [48], but this is an intracellular enzyme that is not involved in lignin degradation. However, its crystal structure has been used as a template to predict the molecular structure of AAO (shown in Fig. 5B, including the FAD cofactor) [46]. It has been shown that AAO is a unique oxidase due both to its spectroscopic characteristics (flavin intermediates and reactivity) and to the wide range of aromatic and aliphatic polyunsaturated primary alcohols (and even aldehydes) that it is able to oxidize [15]. The molecular structure of AAO includes two catalytically active histidines near the N5 of the flavin ring, which might help electron transfer to/from the cofactor by acting as bases in the oxidation of aromatic alcohols (which would proceed via a hydride transfer mechanism) and as acids in the reduction of oxygen to H2O2.
As noted above, laccases were the first ligninolytic enzymes to be investigated, and had been known in plants for many years. Nevertheless, the first molecular structure of a complete fungal laccase was published only in 2002. That year, the crystal structures of the laccases from the basidiomycete T. versicolor, and the ascomycete Melanocarpus albomyces were reported [2,23,37] (Fig. 5C). The first structure of a bacterial laccase was published one year later [11]. A previously reported laccase structure corresponded to an inactive form due to the loss of a copper ion during deglycosylation to obtain suitable crystals for X-ray diffraction [10]. The active site of laccases includes four copper ions. Type-I copper (right sphere in Fig. 5C) acts as electron acceptor from substituted phenols or amines (the typical laccase substrates); and type-II copper, which transfers the electrons to the final acceptor, dioxygen, which is reduced to water. The two type-III coppers act as intermediates in the electron transfer pathway that also includes one cysteine and two histidine protein residues. The molecular environment of laccase type-I copper seems to regulate the redox potential of the enzyme [37]. The fact that laccase can use atmospheric oxygen as the final electron acceptor represents a considerable advantage for industrial and environmental applications compared with peroxidases, which require a continuous supply of H2O2. Taking into account that the advantage of peroxidases is their higher redox potential, engineering the active site of laccases to obtain high redox potential variants would be of considerable biotechnological interest.
Most enzymes involved in wood lignin degradation (a multienzymatic process that includes, among others, peroxidases, oxidases, and laccases acting synergistically) have been identified, and the mechanisms of action of several of them have been established at a considerably precise level. These enzymes, however, cannot penetrate the compact structure of sound wood tissues due to their comparatively large molecular size. Therefore, small chemical oxidizers, including activated oxygen species and enzyme mediators, are probably involved in the initial steps of wood decay.
Acknowledgements. These studies have been partially supported by ENCE (Spain), by Spanish projects AGL2002-393 and BIO2002-1166, by EU projects QLK5-99-1357 and QLK3-99-590, and by an EUFORES(ENCE)-PDT/MEC(Uruguay) grant. Carmen Ascaso (CCMA, CSIC, Madrid) is acknowledged for low-temperature scanning-electron microscopy facilities. Klaus Piontek (ETH, Zurich) is acknowledged for solving the VP crystal structure. Lina Bettucci (Universidad de la República, Montevideo) is acknowledged for an Inocutis jamaicensis strain. M.S acknowledges MEC for a Postdoctoral Fellowship. F.J.R.-D. thanks CSIC for an I3P contract. A.G. and S.C. thank MEC for their "Ramón y Cajal" contracts.
References
1. Banci L, Camarero S, Martínez AT, Martínez MJ, Pérez-Boada M, Pierattelli R, Ruiz-Dueñas FJ (2003) NMR study of Mn(II) binding by the new versatile peroxidase from the white-rot fungus Pleurotus eryngii. J Biol Inorg Chem 8:751-760 [ Links ]
2. Bertrand T, Jolivalt C, Briozzo P, Caminade E, Joly N, Madzak C, Mougin C (2002) Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry 41:7325-7333 [ Links ]
3. Bourbonnais R, Paice MG (1990) Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett 267:99-102 [ Links ]
4. Camarero S, García O, Vidal T, Colom J, del Río JC, Gutiérrez A, Gras JM, Monje R, Martínez MJ, Martínez AT (2004) Efficient bleaching of non-wood high-quality paper pulp using laccase-mediator system. Enz Microb Technol 35:113-120 [ Links ]
5. Camarero S, Ibarra D, Martínez MJ, Martínez AT (2005) Lignin-derived compounds as efficient laccase mediators for decolorization of different types of recalcitrant dyes. Appl Environ Microbiol 71:1775-1784 [ Links ]
6. Camarero S, Sarkar S, Ruiz-Dueñas FJ, Martínez MJ, Martínez AT (1999) Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese-peroxidase and lignin-peroxidase substrate interaction sites. J Biol Chem 274:10324-10330 [ Links ]
7. del Río JC, Gutiérrez A, Martínez AT (2004) Identifying acetylated lignin units in non-wood fibers using pyrolysis-gas chromatography/mass spectrometry. Rapid Commun Mass Spectrom 18:1181-1185 [ Links ]
8. del Río JC, Gutiérrez A, Martínez MJ, Martínez AT (2001) Py-GC/MS study of Eucalyptus globulus wood treated with different fungi. J Anal Appl Pyrol 58/59:441-452 [ Links ]
9. Doyle WA, Blodig W, Veitch NC, Piontek K, Smith AT (1998) Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Biochemistry 37:15097-15105 [ Links ]
10. Ducros V, Brzozowski AM, Wilson KS, Brown SH, Ostergaard P, Schneider P, Yaver DS, Pedersen AH, Davies GJ (1998) Crystal structure of the type-2 Cu depleted laccase from Coprinus cinereus at 2.2 Å resolution. Nature Struct Biol 5:310-316 [ Links ]
11. Enguita FJ, Martins LO, Henriques AO, Carrondo MA (2003) Crystal structure of a bacterial endospore coat component: A laccase with enchanced thermostability properties. J Biol Chem 278:19416-19425 [ Links ]
12. Eriksson K-EL, Blanchette RA, Ander P (1990) Microbial and enzymatic degradation of wood components. Springer-Verlag, Berlin [ Links ]
13. Evans CS, Dutton MV, Guillén F, Veness RG (1994) Enzymes and small molecular mass agents involved with lignocellulose degradation. FEMS Microbiol Rev 13:235-240 [ Links ]
14. Fengel D, Wegener G (1984) Wood: Chemistry, ultrastructure, reactions. De Gruyter, Berlin [ Links ]
15. Ferreira P, Medina M, Guillén F, Martínez MJH, van Berkel WJ, Martínez, AT (2005) Spectral and catalytic properties of aryl-alcohol oxidase, a fungal flavoenzyme acting on polyunsaturated alcohols. Biochem J (in press). Published online April 6, 2005, [doi:10.1042/ BJ20041903] [ Links ]
16. Gold MH, Youngs HL, Gelpke MD (2000) Manganese peroxidase. Met Ions Biol Syst 37:559-586 [ Links ]
17. Guillén F, Gómez-Toribio V, Martínez MJ, Martínez AT (2000) Production of hydroxyl radical by the synergistic action of fungal laccase and aryl alcohol oxidase. Arch Biochem Biophys 383:142-147 [ Links ]
18. Guillén F, Martínez AT, Martínez MJ (1992) Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem 209:603-611 [ Links ]
19. Guillén F, Martínez AT, Martínez MJ, Evans CS (1994) Hydrogen peroxide-producing system of Pleurotus eryngii involving the extracellular enzyme aryl-alcohol oxidase. Appl Microbiol Biotechnol 41:465-470 [ Links ]
20. Guillén F, Martínez MJ, Muñoz C, Martínez AT (1997) Quinone redox cycling in the ligninolytic fungus Pleurotus eryngii leading to extracellular production of superoxide anion radical. Arch Biochem Biophys 339:190-199 [ Links ]
21. Gutiérrez A, Caramelo L, Prieto A, Martínez MJ, Martínez AT (1994) Anisaldehyde production and aryl-alcohol oxidase and dehydrogenase activities in ligninolytic fungi of the genus Pleurotus. Appl Environ Microbiol 60:1783-1788 [ Links ]
22. Gutiérrez A, Martínez AT (1996) Biodegradación de la lignina: Una perspectiva actual. Rev Iberoam Micol 13:18-23 [ Links ]
23. Hakulinen N, Kiiskinen LL, Kruus K, Saloheimo M, Paananen A, Koivula A, Rouvinen J (2002) Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nature Struct Biol 9:601-605 [ Links ]
24. Heinfling A, Ruiz-Dueñas FJ, Martínez MJ, Bergbauer M, Szewzyk U, Martínez AT (1998) A study on reducing substrates of manganese-oxidizing peroxidases from Pleurotus eryngii and Bjerkandera adusta. FEBS Lett 428:141-146 [ Links ]
25. Higuchi T (1997) Biochemistry and molecular biology of wood. Springer Verlag, London [ Links ]
26. Jensen KAJr, Bao W, Kawai S, Srebotnik E, Hammel KE (1996) Manganese-dependent cleavage of nonphenolic lignin structures by Ceriporiopsis subvermispora in the absence of lignin peroxidase. Appl Environ Microbiol 62:3679-3686 [ Links ]
27. Käärik A (1965) The identification of the mycelia of wood-decay fungi by their oxidation reactions with phenolic compounds. Studia Forestalia Suecica 31:3-81 [ Links ]
28. Kersten PJ (1990) Glyoxal oxidase of Phanerochaete chrysosporium: Its characterization and activation by lignin peroxidase. Proc Natl Acad Sci USA 87:2936-2940 [ Links ]
29. Lee KH, Wi SG, Singh AP, Kim YS (2004) Micromorphological characteristics of decayed wood and laccase produced by the brown-rot fungus Coniophora puteana. J Wood Sci 50:281-284 [ Links ]
30. Lin SY, Dence CW (1992) Methods in lignin chemistry. Springer-Verlag, Berlin [ Links ]
31. Martínez AT (2002) Molecular biology and structure-function of lignin-degrading heme peroxidases. Enz Microb Technol 30:425-444 [ Links ]
32. Martínez MJ, Ruiz-Dueñas FJ, Guillén F, Martínez AT (1996) Purification and catalytic properties of two manganese-peroxidase isoenzymes from Pleurotus eryngii. Eur J Biochem 237:424-432 [ Links ]
33. Marzullo L, Cannio R, Giardina P, Santini MT, Sannia G (1995) Veratryl alcohol oxidase from Pleurotus ostreatus participates in lignin biodegradation and prevents polymerization of laccase oxidized substrates. J Biol Chem 270:3823-3827 [ Links ]
34. Mayer AM, Staples RC (2002) Laccase: new functions for an old enzyme. Phytochemistry 60:551-565 [ Links ]
35. Otjen L, Blanchette RA (1986) A discussion of microstructural changes in wood during decomposition by white rot basidiomycetes. Can J Bot 64:905-911 [ Links ]
36. Pérez J, Muñoz-Dorado J, de la Rubia RT, Martínez, J (2002) Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Microbiol 5:53-63 [ Links ]
37. Piontek K, Antorini M, Choinowski T (2002) Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-Å resolution containing a full complement of coppers. J. Biol Chem 277:37663-37669 [ Links ]
38. Piontek K, Glumoff T, Winterhalter K (1993) Low pH crystal structure of glycosylated lignin peroxidase from Phanerochaete chrysosporium at 2.5 Å resolution. FEBS Lett 315:119-124 [ Links ]
39. Poulos TL, Edwards SL, Wariishi H, Gold MH (1993) Crystallographic refinement of lignin peroxidase at 2 Å. J Biol Chem 268:4429-4440 [ Links ]
40. Rodríguez E, Nuero O, Guillén F, Martínez AT, Martínez MJ (2004) Degradation of phenolic and non-phenolic aromatic pollutants by four Pleurotus species: the role of laccase and versatile peroxidase. Soil Biol Biochem 36:909-916 [ Links ]
41. Ruiz-Dueñas FJ, Martínez MJ, Martínez AT (1999) Molecular characterization of a novel peroxidase isolated from the ligninolytic fungus Pleurotus eryngii. Mol Microbiol 31:223-235 [ Links ]
42. Schwarze FWMR, Engels J, Mattheck C (2000) Fungal strategies of decay in trees. Springer, Berlin [ Links ]
43. Shimada M, Takahashi M (1991) Biodegradation of cellulosic materials. In: Hon DNS, Shiraishi N (eds) Wood and cellulosic chemistry. Marcel Dekker, NY, pp 621-663 [ Links ]
44. Sundaramoorthy M, Kishi K, Gold MH, Poulos TL (1994) The crystal structure of manganese peroxidase from Phanerochaete chrysosporium at 2.06-Å resolution. J Biol Chem 269:32759-32767 [ Links ]
45. Sundaramoorthy M, Kishi K, Gold MH, Poulos TL (1997) Crystal structures of substrate binding site mutants of manganese peroxidase. J Biol Chem 272:17574-17580 [ Links ]
46. Varela E, Martínez MJ, Martínez AT (2000) Aryl-alcohol oxidase protein sequence: A comparison with glucose oxidase and other FAD oxidoreductases. Biochim Biophys Acta 1481:202-208 [ Links ]
47. Whittaker MM, Kersten PJ, Nakamura N, Sanders-Loehr J, Schweizer ES, Whittaker JW (1996) Glyoxal oxidase from Phanerochaete chrysosporium is a new radical-copper oxidase. J Biol Chem 271:681-687 [ Links ]
48. Wohlfahrt G, Witt S, Hendle J, Schomburg D, Kalisz HM, Hecht H-J (1999) 1.8 and 1.9 Å resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidase as a basis for modelling substrate complexes. Acta Crystallogr D 55:969-977 [ Links ]
49. Zabel R, Morrell J (1992) Wood microbiology: Decay and its prevention. Academic Press, London [ Links ]