Meu SciELO
Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
Revista de Osteoporosis y Metabolismo Mineral
versão On-line ISSN 2173-2345versão impressa ISSN 1889-836X
Rev Osteoporos Metab Miner vol.8 no.1 Madrid Jan./Mar. 2016
Study of miRNAs expression patterns in osteoporotic bone
Estudio del patrón de expresión de microRNAs en el hueso osteoporótico
García-Giralt N.1, De-Ugarte L.1, Yoskovitz G.1, Güerri R.1,2, Grinberg D.3, Nogués X.1,2, Mellibovsky L.1,2, Balcells S.3 and Díez-Pérez S.1,2
1 IMIM (Instituto Hospital del Mar de Investigaciones Médicas) - Red Temática de Investigación Cooperativa en Envejecimiento y Fragilidad (RETICEF) - ISCIII - Barcelona (España)
2 Departamento de Medicina Interna - Parque de Salud Mar - Universidad Autónoma de Barcelona - Barcelona (España)
3 Departamento de Genética - Universidad de Barcelona - IBUB - Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER) - ISCIII - Barcelona (España)
This paper was supported by the Thematic Network for Cooperative Research in Aging and Fragility (RETICEF; RD12 / 0043/0022), and support FIS PI13 / 00116 (Carlos III Health Institute, Ministry of Science and Innovation) and FEDER.
Work scholarship from the SEIOMM to attend the 36th Congress of the ASBMR (Houston, 2014).
SUMMARY
Objectives: To identify microRNAs (miRNAs) differentially expressed in bone samples with osteoporotic fracture compared with healthy bones.
Methods: Total RNA was extracted from fresh trabecular bone of the femoral neck of women undergoing hip replacement surgery, either because to osteoporotic fracture (n=6) or in the absence of osteoarthritis osteoporosis (based on BMD) (n=6). The samples were hybridized on an array of miRNAs and PCA diagrams and heat map were made. To compare expression levels, >1.5 times and a value p<0.05 Student's T test (corrected for multiple testing) was set as a threshold of significant change.
Results: Both PCA analysis and the heat map showed a samples grouping whether there was fracture or not. 790 were detected miRNAs in bone samples, 82 of which were altered in the osteoporotic samples. After validation in another panel of 6 samples 6 osteoporotic and non-osteoporotic by PCR real time of the most significant miRNAs, and for which there was a test available, the miRNAs, miR-320a and miR-22-3p were confirmed. These two miRNAs were detected in cultures of primary osteoblasts, although they did not maintain the same pattern of expression in total bone samples.
Conclusions: We have shown that there are differences in the expression of miRNAs in samples with osteoporotic fracture. This opens prospects for research and design of new therapies.
Key words: microRNA, bone fracture, osteoporosis, osteoblasts, bone.
RESUMEN
Objetivos: Identificar microRNAs (miRNAs) diferencialmente expresados en muestras óseas con fractura osteoporótica respecto a huesos sanos.
Material y métodos: Se extrajo RNA total a partir de hueso trabecular fresco del cuello femoral de mujeres sometidas a reemplazo de cadera, ya sea debido a fractura osteoporótica (n=6) o por artrosis en ausencia de osteoporosis (según la DMO) (n=6). Las muestras se hibridaron en un array de miRNAs y se realizaron diagramas de PCA y de mapa de calor. Para la comparación de los niveles de expresión, se fijó como significativo un umbral de cambio de >1,5 veces y un valor p<0,05 en la t de Student (corregido para múltiples pruebas).
Resultados: Tanto los análisis de PCA como el mapa de calor mostraron una agrupación de las muestras según si eran de fractura o no. Se detectaron 790 miRNAs en las muestras de hueso, 82 de los cuales estaban alterados en las muestras osteoporóticas. Tras la validación en otro panel de 6 muestras osteoporóticas y 6 no osteoporóticas mediante PCR a tiempo real de los miRNAs más significativos, y para los que existía un ensayo disponible, se confirmaron los miRNAs miR-320a y miR-22-3p.
Estos dos miRNAs se detectaron en cultivos de osteoblastos primarios, aunque no mantenían el mismo patrón de expresión que en las muestras de hueso total.
Conclusiones: Hemos demostrado que existen diferencias en la expresión de miRNAs en muestras con fractura osteoporótica, lo que abre nuevas perspectivas para la investigación y diseño de nuevas terapias.
Palabras clave: microRNA, fractura ósea, osteoporosis, osteoblastos, hueso.
Introduction
MicroRNAs (miRNAs) have been linked to a variety of processes, such as cellular proliferation, differentiation and apoptosis. Deregulation of any of these processes could lead to pathological disorders, some as severe as cancer [1].
MicroRNAs are small (18-24nt), non-coded RNAs, that negatively regulate gene expression by binding to the 3'-UTRs of the target miRNAs sequences. In bone tissue, miRNAs have been described as key factors in regulating the formation, remodeling and homeostasis of the bone [2,3]. Furthermore, several studies have shown that miRNAs are involved in controlling the differentiation and function of bone cells [4].
Thus the identification of such miRNAs could be a tool to develop therapies to promote bone formation or inhibit bone resorption and so act on bone diseases.
In the field of osteoporosis there are very few studies concerning miRNA involvement in its pathophysiology. Li, et al. [5] described a mutation in the pre-miR-2861 that blocked miR-2861expression, causing primary osteoporosis in two adolescent relations. In another study, three polymorphisms in the target sequences for miR-146a and miR-146b in the FGF2 gene is genetically associated with bone mineral density (BMD) of the femoral neck [6]. There have also been several studies which attempted to identify miRNAs with an altered expression pattern associated with osteoporotic fracture to find disease biomarkers [7-9]. All of these studies found different miRNAs proving once again the complexity of osteoporosis fracture.
However, the sample type, the conditions of collection and handling process as well as the characteristics of the study population may influence the final result. In fact, a variety of expression profiles of miRNAs have been found depending on the cell type studied within the osteoblast lineage (proliferation, differentiation, mineralization), if subjected to any treatment (hormones, cytokines) or species of origin [10,11]. Thus, it is difficult to compare studies, although any input in the field is a step closer to understanding the pathophysiology of the fracture.
The aim of this study was to identify miRNAs with altered expression in osteoporotic bone, using an experimental methodology as close as possible to physiological conditions. For this purpose, the trabecular bone obtained from patients with a recent osteoporotic fracture compared to non-osteoporotic bone samples were analyzed. A study of microarray hybridization was performed using fresh whole bone tissue to detect all miRNAs expressed in these samples.
Material and methods
Preparation of the bone samples
The trabecular bone of the femoral neck was obtained from postmenopausal women undergoing hip replacement, either osteoporotic fracture (OP) (n=6) or osteoarthritis (n=6). Samples in the absence of osteoarthritic osteoporosis were determined by measuring BMD and considered as the control group. To validate the results of the array, six additional samples of the trabecular bone of the femoral neck with fracture were obtained and 6 samples without osteoporosis. Of the latter samples, primary osteoblasts (HOB) were also obtained. No patient presented a history of metabolic or endocrine disease, chronic renal failure, chronic liver disease, cancer, Paget's disease, malabsorption syndrome, treatment of hormone replacement therapy, or medication agents or oral anabolic steroids, anti-epileptic drugs, lithium heparin or warfarin. Written informed consent was obtained in accordance with the regulations of the Ethics Committee of the MAR Health Park Clinical Research Center, which approved the study.
Cultivation of primary osteoblasts
For the cultivation of osteoblasts from the bone samples of the validation phase, small fragments of trabecular bone were obtained and placed in culture plates of 140 mm. They were incubated with DMEM (Dulbecco's Modified Eagle Medium) culture medium supplemented with FBS (Fetal Bovine Serum) 10% penicillin/streptomycin 1%, 0.4% fungizone and 100 μg/ml ascorbic acid. The plates were trypsinized after about three weeks just before the junction for RNA extraction.
RNA extraction
For RNA extraction, total bone tissue, fresh samples of trabecular bone were cut into small pieces, washed three times in phosphate buffered saline (PBS), and stored at -80 until use.
The RNA extraction of both total bone and HOB was performed using miRNeasy Mini Kit (Qiagen) following the manufacturer's instructions. For primary osteoblasts, the RNeasy MinElute Cleanup (Qiagen) was also used to obtain the fraction enriched miRNAs. The concentration of purified RNA was analyzed in a spectrophotometer (Nanodrop, Thermo Fisher Scientific Inc).
Microarray microRNAs of total bone samples and data analysis
The microarray and data analysis were performed on the Exiqon (Denmark) platform platform. The quality of total RNA was verified by the Agilent 2100 Bioanalyzer and 250 ng of RNA, both as reference samples, were labeled with fluorescent signals HY3™ and HY5™, respectively, using the miRCURY LNA™ microRNA Hi-Power Labeling Kit, HY3™/HY5™ (Exiqon, Denmark) following the procedure described by the manufacturer. The RNA samples, labeled HY3™, and the sample labeled reference HY5™ were mixed equally and hybridized to the array of miRCURY LNA™ microRNA (Exiqon, Denmark) containing capture probes for all human, mouse or rat miRNAs, entered into the miRBase 18.0. Hybridization was carried out following the array instructions using a Tecan HS4800 hybridization station™ (Tecan, Austria). After hybridization, the results were scanned and stored in an environment free of ozone (<2.0 ppb ozone) to prevent the extinction of the fluorescent markers. Scanning was carried out using the Agilent Microarray Scanner System G2565BA (Agilent Technologies, Inc., USA). Image analysis was performed using ImaGene® 9 (Software Analysis miRCURY LNA™ microRNA, Exiqon, Denmark). Signals were quantified (Normexp With offset value 10, see Ritchie, et al. [12]) and normalized using the global regression algorithm Lowess (locally weighted scatterplot smoothing). After normalization, unsupervised and supervised data analysis took place. Diagrams of principal component analysis (PCA) and heat map showing performed unsupervised hierarchical clustering. The expression levels were compared using the Student t test. The significance threshold was established in the change log (logC) >1.5 times and a value of p<0.05.
Validation of miRNAs differentially expressed between the OP group and the control group
The quantification of the expression of miRNAs in samples of whole blood was performed by real-time PCR (qPCR) on the Exiqon (Denmark) platform. 10 ng of RNA were transcribed to cDNA using the kit miRCURY LNA™ Universal RT microRNA PCR, polyadenylation and cDNA synthesis according to the instructions. The cDNA was diluted 100-fold and the expression of each of the miRNAs was quantified by qPCR using a custom panel of specific primers (panel custom pick & mix, Exiqon) and ExiLENT SYBR® Green mastermix. Negative controls were run in parallel with the samples. Amplification was performed on a LightCycler 480 Real-Time PCR System (Roche) in 384-well plates. Amplification curves were analyzed using the software Roche LC Cq for determining the value (the second derivative method). Differences in expression levels between the two groups was calculated as 2^ddCq. Amplification efficiency was calculated using algorithms similar to LinReg software. In the analysis of the test data, only 3 Ct detected below the negative control and Ct <37 were included. Normalization was performed based on the test average detected in all samples, as it is proven to be the best method for normalization of qPCR studies involving numerous tests [13]. For the present study, this included 11 trials. The stability of the mean of 11 miRNAs was higher than any miRNA alone in the measured data set by NormFinder software [14].
The formula used to calculate normalized values Cq (DCQ) is:
Standard Cq = Cq Media - Cq trial (sample)
Finally, the quality control of the data was performed as well as unsupervised data analysis, Student t and Wilcoxon test for comparison between groups (p<0.05 was considered significant).
Validation primary osteoblasts
To prepare cDNA, 1 µg of RNA was retrotranscribed from each sample using the miScript II RT kit (Qiagen).
The expression of miRNAs was quantified by qPCR using the miScript SYBR Green PCR Kit using the sequence of the mature miRNA as the first. The cDNA was diluted at a ratio of 1/5 and 0.5 µl of sample per well was used, following the protocol described by the supplier. Amplification was performed in the "Flex QuantStudio 12K Real-Time PCR" system in 384-well plates, and the data were analyzed with "expression suite" software. Expression was analyzed by RQ relative quantification using the method of the second derivative (DDCt). The U6 snRNA was used as endogenous control to normalize the samples. Each experiment was done in triplicate. To compare the statistical differences between the groups with and without osteoporosis, Mann-Whitney nonparametric statistical test was carried out using SPSS version 12.0 for Windows.
Bioinformatic analysis of miRNAs validated
For studying target genes differentially expressed miRNAs was used following programs: PicTar (http://pictar.mdc-berlin.de) TargetScan Human (http://www.targetscan.org) miRDB (http://mirdb.org), Miranda (http://www.microrna.org) DIANA-TarBase (http://diana.imisathena-innovation.gr) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw). TARGET-mirPath computer based tool web (11) to identify potentially altered molecular pathways and the intersection of the miRNAs differentially expressed in the fractured bone was used. Information about protein function was obtained in the UniProtKB database (http://www.uniprot.org).
Results
Description of the study patients
Anthropometric characteristics, both of patients whose bone samples were used for expression array, as well as those used to perform validations array results are shown in Table 1. There were no differences in age and body mass index between the two groups of patients (Mann-Whitney test).
Unsupervised analysis of the expression array
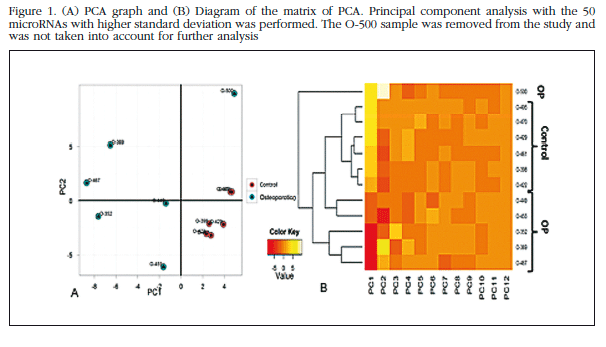
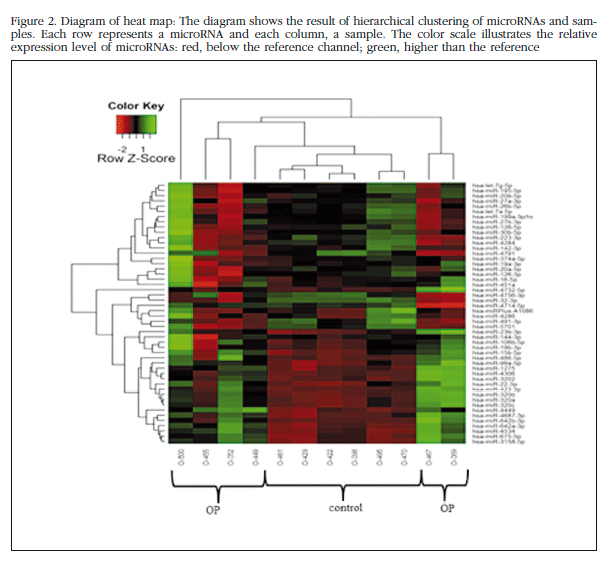
Each trabecular bone sample of all the patients in the study bone was analyzed individually in the microarray miRNAs, and unsupervised analysis of the results was carried out based on the expression profile in order to identify variation patterns related to biological and technical factors. Principal component analysis (PCA) was performed which included 50 miRNAs with greater expression variation between samples to get an overview of the clustering of the samples according to their variance (Figure 1). The group of non-osteoporotic samples (control group) showed a much more homogeneous profile than the osteoporotic samples. The O-500 sample obtained from a patient with osteoporosis, was considered atypical and excluded from the analysis. The heat map diagram corroborates the PCA results with a clear grouping of the control samples and a sparse array of osteoporotic samples (Figure 2).
Comparison of expression of microRNAs between OP and controls in total bone samples
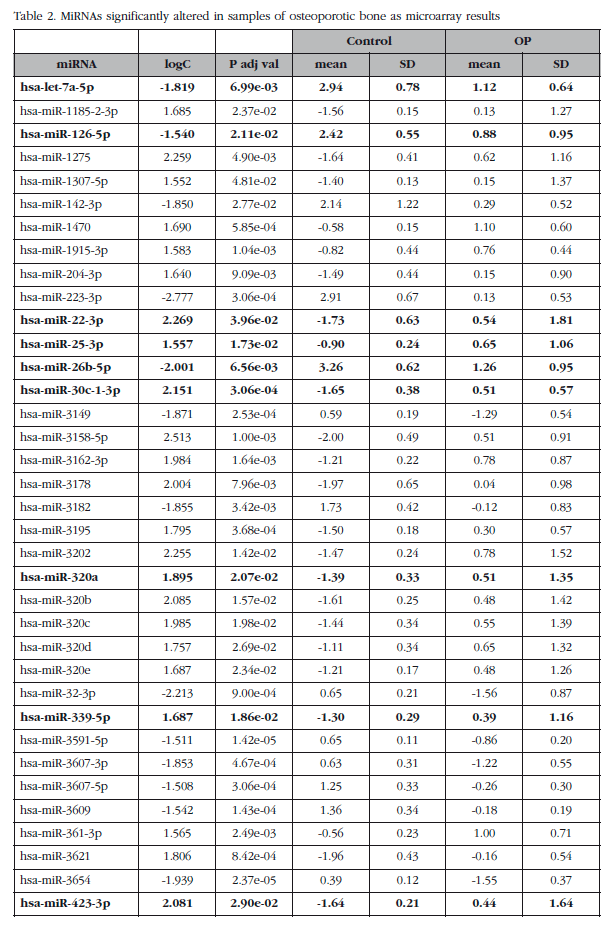
The mean expression levels of miRNAs were compared between groups with and without osteoporosis, excluding the O-500 samples. This analysis identified a subset of 82 miRNAs (miRNAs analyzed about 1,932) whose absolute value of the logarithm of the ratio was greater than 1.5 and with an adjusted p value less than 0.05. Seven of these miRNAs corresponded to small nucleolar RNAs, C/D box (SNORD), three to virus and one was a miRPlus™ property Exiqon sequence is not recorded in the miRBase data, and were excluded from the validation phase.
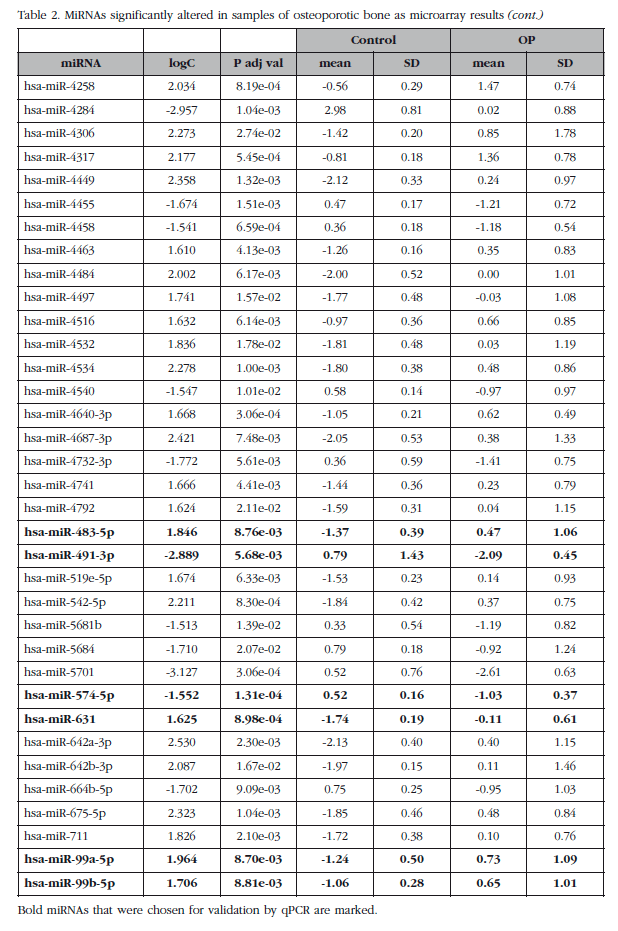
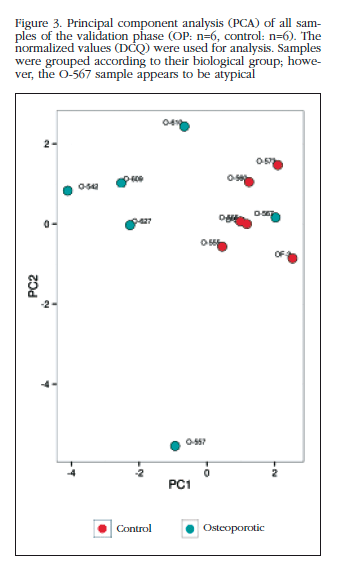
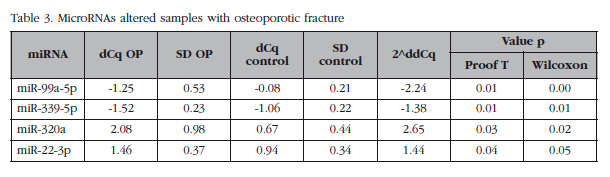
Of those remaining (Table 2), 15 hsa-miRNAs were chosen with the best significance values and for which Exiqon available probes for validation by qPCR: let-7a-5p, miR-126-5p, miR-30c- 1-3p, miR-22-3p, miR-25-3p, miR-26b-5p, miR-339-5p, miR-423-3p, miR-320a, miR-483-5p, miR-491-3p, miR-574-5p, miR-631, miR-99a-5p and miR-99b-5p. The PCA diagram of individual qPCR results showed clustering of the sample results to be very similar to the array expression, confirming the different biological source of the two sample groups (Figure 3). An osteoporotic sample (O-567) was located in the control group and was excluded from the analysis. After statistical analysis four miRNAs; miR-320a, miR-99a-5p, miR-339-5p and miR-22-3p showed significant differences between groups with osteoporosis and control (Table 3). However, the miRNAs miR-99a-5p and miR-339-5p were found to be overexpressed in OP samples results array expression, while they were under-expressed in the OP samples validation phase. These contradictory results suggest a role for these miRNAs unrelated to osteoporotic disease and they were discarded for in silico study.
Comparison of expression of microRNAs between OP and controls crops Hob
For each of the whole blood samples, primary osteoblasts were obtained. These were analyzed by qPCR expression of the miRNAs previously validated in bone tissue samples. MiRNAs miR-99a-5p and miR-339-5p were not detected in osteoblastic cells, while the miRNAs miR-320a and miR-22-3p were expressed in the HObs, although neither showed significant differences between the two biological groups.
Prediction of target genes and analysis of signaling pathways validated miRNAs
A comprehensive computer analysis by six different programs allowed us to predict potential target genes for the validated miRNAs. MiR-320a can regulate genes involved in cell proliferation (KRAS, PDGFD), inhibition of apoptosis (MCL1), signal transduction (MAPK1, SOS2, PTEN), regulation of gene expression (RUNX2, PPARGC1A, SP1, CAMTA1, ESRRG) receptors, growth factors, hormones and cytokines (NPR1, BMPR1A, AR, IGF1R, ESR1), etc. Moreover, as validated targets are the CTNNB1, TFRC and POLR3D genes. This miRNA is involved in 44 of the Encyclopedia of Genes and Genomes Kyoto (KEGG) tracks, according to predictions of the DIANA-mirPath program where the prostate cancer path is the most significant (1,105e-12), followed by endometrial cancer (2,261e-10), the mTOR signaling pathway (7,132e-08), and the PI3K-Akt signaling pathway (3,914e-07).
The miR-22-3p has many validated target genes, some of them very important in the regulation of bone metabolism. Among them are the ESR1, the PRKACA (required for adipogenic differentiation and inhibition of osteoblast differentiation), HDAC4, SP1, BMP7 and CDK6 (BMP2 signaling antagonist). The main signaling pathway KEGG This miRNA is endocytosis (4,21e-05) but also acts in important signaling pathways such as p53 (p=0.003) and MAPK (p=0.003).
Diana software tools using the validated genes (according to the Tarbase data base) shows the endocytosis pathway as the route of intersection of the two miRNAs as they share the TFRC target gene (transferrin receptor). Furthermore, if the intersection is evaluated considering the predicted target genes according to the MicroT_CDS data base, the signaling pathways are the most significant for prostate cancer and mTOR where the two miRNAs share AKT3, PTEN and IGFR1 target genes.
Discussion
This study focuses on identifying miRNAs with altered expression in osteoporotic bone. To achieve this, fresh trabecular bone was collected from patients with or without osteoporotic fracture, with homogeneous anthropometric parameters such as age, BMI and gender. We excluded patients with disorders that affect bone remodeling. Samples from osteoporotic fracture showed a clear involvement in the expression pattern of miRNAs, demonstrating that epigenetic regulation is altered in pathological bone. These results may provide a better understanding of bone biology into those who suffer an osteoporotic fracture. Also, they help identify molecules that can be used as therapeutic targets.
Unsupervised analysis of the results of miRNAs array expression from total bone samples showed that non-osteoporotic samples (control) were grouped generating a biological cluster. Furthermore, osteoporotic bone clearly differed from the control samples, showing a more dispersed distribution, suggesting that the pathophysiology of osteoporotic disease and, ultimately, bone fracture, have a heterogeneous etiology. Working with fresh human samples generates greater variability to explore established cell lines or animal models, making it difficult to see significant differences between groups. Still, this is closer to the pathophysiological situation scenario. In this regard, our study is based on samples of human bones obtained with minimal laboratory handling, which allows us to get a similar situation to in vivo conditions.
In the analysis of differential expression between osteoporotic and control samples, 82 miRNAs that reached significance levels with an absolute value of the logarithm of the ratio greater than 1.5 times found. Of these, 15 were tested by qPCR in a new set of samples and the miRNAs miR-99a-5p, miR-339-5p, miR-320a and miR-22-3p were finally validated. These miRNAs were evaluated in primary osteoblasts (HOB) in culture, cells from osteoporotic comparing samples versus cells from non-osteoporotic samples. MiRNAs miR-99a-5p and miR-339-5p were not detected in the HOB, suggesting another cell type as the source of these two miRNAs. Moreover, these miRNAs were found overexpressed in array osteoporotic samples whereas they were under-expressed in the validation phase. This would seem to discard these miRNAs' link to osteoporosis. MiRNAs, miR-320a and miR-22-3p expressed in the Hob, although no differences in expression levels between sample groups were observed. This could be due to the artificial conditions of the in vitro cell cultures that affect the expression of microRNAs, especially those involved in the regulation of important cellular functions such as proliferation and differentiation.
This shows us once again that, although the use of cell cultures, both in established and primary lines, can help researchers understand certain cellular processes. The results may differ from the actual conditions that would occur in the original tissue within the physiological environment.
Furthermore, the use of fresh tissue not manipulated for closer physiological conditions can also provide information such dissimilar results as demonstrated in studies by Seeliger, et al. [7] and Garmilla-Ezquerra, et al. [8]. These studies also conducted a microarray expression from whole bone samples comparing osteoporotic versus non osteoporotic samples very similar to ours, but with different findings. Note that these two earlier studies have several features that can explain these discrepancies. These include the variety of commercial arrays used and the sample size. Furthermore, these biological studies comparing groups with anthropometric characteristics or inhomogeneous clinical features, such as age, gender, body mass index and endocrine disorders (eg. diabetes mellitus), essential for the regulation of bone metabolism. Therefore, the miRNAs identified in these studies could also be linked to other external parameters, whereas in our study, the absence of concomitant diseases, and similar features between the two groups, allows us to approach in a more reliable way what happens in osteoporotic fracture.
Therefore, a major effort in our work is extremely careful control of potential confusing characteristics between cases and controls for age, sex, body mass index and metabolic diseases associated with aging. These strict inclusion criteria considerably restrict our sample size compared to similar works, where the samples are from patients with heterogeneous characteristics. Another limitation of our study is that non-osteoporotic samples used as controls from patients with osteoarthritis and, therefore, cannot rule out other bone abnormalities. For ethical reasons collecting bone it is not allowed from healthy individuals. However, in an attempt to minimize this potential problem, the sample was obtained from bone located as far as possible the arthritic lesion.
MiR-320a is conserved in human, mouse, rat and cow. This miRNA sequence is located within the basal promoter POLR3D gene, which in turn is one of their target genes and their expresión [15] silencing. Furthermore, it has been observed that miR-320a is involved in regulating osteoblast function as it has as target the CTNNB1 gene (encoding B-catenin) [16] and RUNX2 [17].
Moreover, miR-22-3p is a serum miRNA that was previously associated with osteoporotic fracture [9] and has been implicated in the osteogenic differentiation [18].
Our study results, along with other work in this field, provide an important understanding of bone biology and the involvement of miRNA in the pathology of osteoporosis.
Conclusions
We have identified two miRNAs that are overexpressed in trabecular bone samples from patients with osteoporosis. The expression of both miRNAs is detected in primary osteoblasts, although this overexpression has not been observed in cultures from osteoporotic samples. It is not known whether the alteration of these miRNAs is a cause or effect of the disease and its relationship with osteoporotic fracture. However, these miRNAs could offer promising potential for designing new drugs for osteoporosis.
Conflict of interests
The authors declare no conflict of interest in relation to this work.
![]() Correspondence:
Correspondence:
Natalia García Giralt
c/Dr. Aigüader, 88
08003 Barcelona (España)
Correo electrónico: ngarcia@imim.es
Date of receipt: 20/10/2015
Date of acceptance: 10/12/2015
Bibliography
1. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol 2007;302:1-12. [ Links ]
2. Taipaleenmaki H, Bjerre Hokland L, Chen L, Kauppinen S, Kassem M. Mechanisms in endocrinology: micro-RNAs: targets for enhancing osteoblast differentiation and bone formation. Eur J Endocrinol 2012;166:359-71. [ Links ]
3. Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 2012;8:212-27. [ Links ]
4. van Wijnen AJ, van de Peppel J, van Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, et al. MicroRNA Functions in Osteogenesis and Dysfunctions in Osteoporosis. Curr Osteoporos Rep 2013;11:72-82. [ Links ]
5. Li H, Xie H, Liu W, Hu R, Huang B, Tan YF, et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J Clin Invest 2009;119:3666-77. [ Links ]
6. Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res 2011;26:72-8. [ Links ]
7. Seeliger C, Karpinski K, Haug A, Vester H, Schmitt A, Bauer J, et al. Five Freely Circulating miRNAs and Bone Tissue miRNAs are Associated with Osteoporotic Fractures. J Bone Miner Res 2014;29:1718-28. [ Links ]
8. Garmilla-Ezquerra P, Sanudo C, Delgado-Calle J, Perez-Nunez MI, Sumillera M, Riancho JA. Analysis of the Bone MicroRNome in Osteoporotic Fractures. Calcif Tissue Int 2015;96:30-7. [ Links ]
9. Weilner S, Skalicky S, Salzer B, Keider V, Wagner M, Hildner F, et al. Differentially circulating miRNAs after recent osteoporotic fractures can influence osteogenic differentiation. Bone 2015;79:43-51. [ Links ]
10. Guo L, Zhao RC, Wu Y. The role of microRNAs in self-renewal and differentiation of mesenchymal stem cells. Exp Hematol 2011;39:608-16. [ Links ]
11. Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther 2011;13:220. [ Links ]
12. Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics 2007;23:2700-7. [ Links ]
13. Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 2009;10:R64. [ Links ]
14. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 2004;64:5245-50. [ Links ]
15. Kim DH, Saetrom P, Snove O Jr., Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A 2008;105:16230-5. [ Links ]
16. Sun JY, Huang Y, Li JP, Zhang X, Wang L, Meng YL, et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting beta-catenin. Biochem Biophys Res Commun 2012;420:787-92. [ Links ]
17. Yu F, Cui Y, Zhou X, Zhang X, Han J. Osteogenic differentiation of human ligament fibroblasts induced by conditioned medium of osteoclast-like cells. Biosci Trends 2011;5:46-51. [ Links ]
18. Trompeter HI, Dreesen J, Hermann E, Iwaniuk KM, Hafner M, Renwick N, et al. MicroRNAs miR-26a, miR-26b, and miR-29b accelerate osteogenic differentiation of unrestricted somatic stem cells from human cord blood. BMC Genomics 2013;14:111. [ Links ]











 texto em
texto em