INTRODUCTION
NAD(P)H: quinone oxidoreductase 1 (NQO1) is a ubiquitous cytosolic flavoenzyme that catalyzes two- electron reduction of various endogenous and exogenous quinones by using flavin adenine dinucleotide (FAD) as a cofactor. NQO1 is largely considered as a detoxifying enzyme which is a key catalyst of reactive quinones and quinone-imines to their less reactive and less toxic hydroquinones forms [1]. In addition to creating substrates for phase II conjugation reactions and promoting excretion, this two-electron reduction route avert the potentially toxic semiquinone radical intermediates. Hydroquinones are not redox-stable and in some condition metabolism by NQO1 yields an additional active sutrates [2]. NQO1 expression has been identified in many human tissues, and its rate of expression controlled by the antioxidant response element (ARE) both in basal and during oxidative stress environments [3]. NQO1 gene contains ARE sequence in its promoter region and has been shown to be regulated by nuclear factor (erythroid-derived 2)-like 2 (Nrf2) [4]. Nrf2 belongs to basic region-leucine zipper family, a member of the Cap 'n' Collar of transcriptional proteins that binds to ARE leading to induction and activation of various antioxidant and cytoprotective genes. Nrf2 bind ARE and control the induction and expression of NQO1 gene [5]. While, Nrf2 knockout exhibited reduced constitutive expression of NQO1 and weakens its induction [3].
The expression NQO1 gene is induced together with additional detoxifying enzyme genes in response to xenobiotics, antioxidants, electrophiles, and heavy metals [6]. Several dietary phytochemicals have been recognized as potential chemopreventive agents. Various fruits and vegetables such as; blueberries, broccoli and cacao beans, are among the most defensive specifically due to an excess of active molecules such as polyphenols, isothiocyanates, and flavonoids [7]. Upon inside cells, these phytochemicals can directly scavenge free radicals and aggravate electrophilic stress signals that trigger proteins related to assorted cellular signalling pathways [8].
THE HUMAN NQO1 GENE EXPRESSION
NAD(P)H:quinone oxidoreductase 1 (NQO1) (originally known as DT-diaphorase,) is a flavoenzyme that plays a crucial role in protecting cells from a broad range of substrates by catalyzing two- and four-electron reductions of endogenous and exogenous quinones to their hydroquinone forms [9]. The two- and four-electron reductions action catalyzed by NQO1 are of importance to the cell by preventing redox cycling, which results in generation of free radicals; thus, NQO1 help to protect the cell from oxidative stresses [10]. According to toxicological classification, NQO1 is regarded as a xenobiotic‐metabolizing phase II enzyme, together with oxidases, reductases and hydrolases that help in introducing a functional group into xenobiotics [11]. NQO1 serve as a homodimer with one flavin adenine dinucleotide (FAD) bound per monomer. This enzyme uses reduced form of pyridine nucleotide cofactors nicotinamide adenine dinucleotide (NADH) or NADPH to catalyze the two-electron reduction of many quinone substrates [12].
NQO1 is localized generally in the cytosol but it also identified at low levels in the nucleus. In human tissues, high levels of NQO1 have been found in diverse epithelial cells, vascular endothelium and adipocytes [13, 14]. Unlike most other mammals, humans do not express NQO1 in normal liver cells but its expression is demonstrated in pre-neoplastic tumors and liver cancers [15, 16, 17]. NQO1 is abundant in many human solid tumours such as; colon, pancreas, lung and breast tumours [18, 19]. The human NQO1 gene (also referred as DIA4) is situated on chromosome 16q22.1; the gene spans around 17 kb and has six exons [20]. There are three of four polyadenylation positions in exon 6 can result in transcripts of 1.2 kb, 1.7 kb, and 2.7 kb in length [21]. NQO1 is coordinately induced by cancer chemopreventive agents with certain phase II drug-conjugating enzymes including glutathione S-transferase (GST) and UDP-glucuronosyl transferase (UGT) isoenzymes [22].
THE CATALYTIC ACTIONS OF NQO1
Ernster et al. in 1958 was initially described the catalytic actions of NQO1 [23]. NQO1 gene is an obligate two-electron reductase of quinones that catalysis reduction of a broad range of substrates by using either NADH or NADPH as reducing cofactors and by its efficient inhibition by dicumarol. It is frequently a cytosolic enzyme (around 90%) and exists as a homodimer with one molecule of FAD per monomer. Over-expression of NQO1, as in oxidative stress status, is commonly observed across many species and offers the cell with protection strategies, possibly by its ability to diminish reactive quinones and quinone-imines to its less toxic and less reactive hydroquinones forms. In addition, two electron reductions bypasses reactive and toxic semiquinone construction and therefore inhibits the production of reactive oxygen species derived from the interaction of the semiquinone with oxygen molecule [24]. The oxidoreductase works through ‘ping-pong' kinetic mechanism where the reduced pyridine nucleotide binds to the active site, reduces the flavin co-factor to FADH2, and is then the oxidized pyridine nucleotide is released before binding of the substrate and thereby total reduction by hydride transfer occur [25]. NQO1 shows activity towards many reactive species including quinones, quinone-imines, glutathionyl-substituted, methylene blue, dichlorophenolindolphenol, naphthoquinones, azo and nitro compounds, demonstrates its significance as a chemoprotective enzyme [26, 27, 28]. In addition to a single two-electron reduction, NQO1 is also able of catalyze four-electron reduction of azo dyes and nitro substances [29, 30]. Using heterodimers of NQO1, it was recognized that the NQO1 subunits work independently in metabolizing two electron substrates and in a dependent way with four-electron substrates [31].
EVIDENCE FOR THE ANTIOXIDANT ACTIVITY OF NQO1
Several previous studies have proposed that NQO1 may involve in the unique antioxidant role, primarily by endogenous quinones reduction which defend cellular membranes against lipid peroxidation. NQO1 protects the cell from unwanted oxidation by maintaining the reduced form of endogenous antioxidants. Mostly, both ubiquinone (co-enzyme Q) and α- tocopherol quinone, two crucial lipid-soluble antioxidants, are substrates for NQO1 in vitro [32, 33]. Beside to its catalytic action in reducing quinones, NQO1 directly able to scavenge superoxide, and this activity could offer an extra protection and therefore efficient in tissues with low level of SOD enzyme expression. Thus, in endothelial cells, where expression of NQO1 is high and that of SOD is relatively low, expression of NQO1 was demonstrated to correlate with increasing superoxide scavenging activity, while its suppression result in a reduction in superoxide scavenging action [34].
Previous studies on rat liver have described that NQO1 can stimulate the reduction of ubiquinone analogues (coenzyme Q) to their ubiquinol forms in liposomes and rat hepatocytes [32, 34]. The decreased rate of coenzyme Q derivatives was dependent upon chain length, with molecules containing longer chains being less efficiently reduced. In these studies, it was established that the ubiquinol formed following the reduction by NQO1 was an effective antioxidant that can protect membrane phospholipids against lipid peroxidation. Beside its role in the electron transport chain, coenzyme Q also works as an antioxidant enzyme and is found in most of cell membranes in its reduced (ubiquinol) form [35]. Additionally, NQO1 elaborated in the metabolism of vitamin E (α-tocopherol) derivatives. α-tocopherol-quinone, a product of α-tocopherol oxidation has been demonstrated to have antioxidant activities following reduction to α- tocopherol hydroquinone [36]. In previous human NQO1 study, it was revealed that this enzyme is active against α-tocopherol-quinone, promoting its reduction to α-tocopherol hydroquinone, and this effect protects against damage to the cell membrane. Furthermore, another study have also showing that Chinese hamster ovary cells transfected with human NQO1 produced α-tocopherol hydroquinone in a high level and were more resistant to lipid peroxidation than NQO1 knockout cells [37]. The protective role of NQO1 against oxidation is also confirmed in humans' studies, in which NQO1 protein is expressed in several tissues such as; epithelial cells of breast, lung and colon, offering a significant level of protection against oxidative stresses. The high levels of NQO1 indicate that NQO1 may serve primarily in an antioxidant tool in these cells. However, biochemical and immunological-based experiments have not succeeded to identify substantial levels of NQO1 expression in human liver [38, 39]. This suggests that in human, distinct from other species, NQO1 does not evolve in a master role in hepatic xenobiotic metabolism.
REGULATION OF NQO1, AND OTHER ARE-DRIVEN GENES, BY BZIP PROTEINS
The ability of various dietary phytochemical to up-regulate the level of NQO1 is arbitrated through the Nrf2-Keap1/ARE system pathway, by regulating the expression of a several phase II detoxifying enzyme genes. Nrf2-Keap1 system helps in protection of mammalian cells against many electrophilic and xenobiotics. The Nrf2-Keap1/ARE pathway is considered as the master regulator of cytoprotective responses to xenobiotics and oxidative stresses [40]. Three main cellular components are of importance for the regulation of Nrf2-Keap1/ARE system pathway transcription, that is; (i) Nrf2, a basic leucine zipper transcription factor, which is a member of the cap 'n' Collar family of regulatory transcriptional proteins that also includes Nrf1, Nrf3, NF-E2, Bach1, and Bach2 [35]. Once activated, Nrf2 binds to ARE transcriptional sites in the promoter regions of several cytoprotective and detoxifying genes, leading to the upregulation of numerous target phase II detoxifying genes that boost cellular detoxification processes and antioxidant potential [41]; (ii)as an adaptor for Cul3-based E3 ligase [42, 43, 44]; and (iii) ARE (Antioxidant response elements), a cis-acting enhancer element presented in the promoter region of various genes encoding phase II detoxification enzymes and have the consensus: TGAG/CNNNGC [45, 46]. For NQO1gene, the precise sequence was reported by John Hayes et al. who revealed that particular nucleotides formerly thought to be redundant in the ARE function have important roles, while others that were formerly considered crucial, were unnecessary [47].
Numerous reports have been conducted in order to prove molecular mechanisms responsible for stimulating Nrf2-Keap1/ARE system pathway. Under basic physiological settings, Keap1 physically sequestered inactive Nrf2 protein in the cytoplasm, thereby blocking its movement to the nucleus (Figure 1). In presence of oxidative stress, Nrf2 translocate into the nucleus, thus initiate Nrf2-ARE transcriptional activation [48]. Numerous models have been reported for the mechanism of regulation of the Keap1/Nrf2/ARE pathway [49]. The most commonly recognized model proposed that Nrf2 inducers alter highly reactive cysteine residues of the sensor Keap1. As consequence, cysteine residues will form protein-protein cross links after exposure to electrophilic or oxidative stress, disturbing Nrf2/Keap1 stabilization and subsequent escaping of Nrf2 [50].
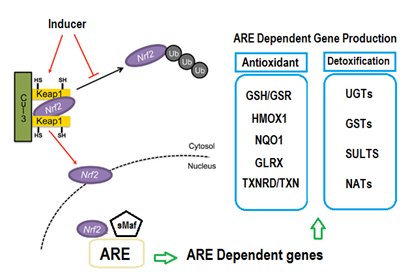
Figure 1. General scheme for the induction of cytoprotective genes through the Nrf2/Keap1/ARE-signaling pathway.
In mammalian tissues, NQO1 is considered as one of the remarkable permanently and efficiently inducible genes among other relevant cytoprotective genes. This early finding has been recurrently agreed by comprehensive gene expression profiling in various systems that used both Nrf2-Keap1/ARE system inducers, and Keap1 knockout genetic methods [51]. NQO1 is significantly expressed in several tumour tissues including lung, liver, colon, breast and pancreatic tissues, and its expression induced in response to a variety of xenobiotics, antioxidants, oxidants, and heavy metals [52]. Mutational experiments in the NQO1 gene promoter helped in identifying the core ARE sequence. ARE is essentially needed for expression and constant activation of NQO1 and other phase II metabolizing genes [53]. Nrf2 bind ARE and upregulate the expression and activation of NQO1 gene. While, knockout of Nrf2 showed reduction in the basal expression of NQO1 and impairs its initiation [3]. In addition, treatment with BHA, known Nrf2 activator, increases hepatic and intestinal NQO1 levels in wild type, but not in Nrf2 knockout mice [54]. All of these data confirm a role for Nrf2 in the expression of NQO1 in various tissues.
POLYMORPHINS IN NQO1
Two single nucleotide polymorphisms (SNPs) have been identified in the human NQO1 gene. The most common variant of NQO1 is a C to T replacement at nucleotide location 609 of the NQO1 cDNA (rs1800566), also referred to as NQO1*2. This nucleotide variation responsible for the alteration of proline to serine amino acid at position 187 (P187S) that is accompanied with a decrease of enzyme action due to variability of the protein product [55]. The variant NQO1*2 protein is extremely unstable, and is promptly ubiquitinated by the proteasome [56]. Thus, the activity of the homozygous variant genotype (NQO1*2/*2) enzyme is substantially undetectable, whereas NQO1*1/*2 heterozygotes exhibit activities intermediate between the homozygous polymorphism genotype and wild type (NQO1*1/*1) [57]. The occurrence of this variant differs broadly by race and a relation were described between the presence of variant alleles in lung and urological cancers [58, 59]. The interethnic variances in this gene were documented to ranges from 16% - 49% in Caucasians to Chinese populations, respectively and the frequency of NQO1*2/*2 homozygosity is reported to range between 1.5% - 20.3% in various ethnic groups [60].
Additional genetic alteration of NQO1 is a substitution of single nucleotide from C to T at codon 465 (rs4986998), also known as NQO1*3, which is responsible on the alteration of the amino acid at codon 139 from arginine to tryptophan (R139W). This polymorphism is similar to the wild type NQO1*1, results in substitutional messenger RNA splice sites that can result in a deletion of exon 4 and produce a protein deficient in quinone binding site for which enzyme activity differs according to the substrate [61, 62, 63]. The NQO1*3 SNP frequency is generally low and ranges from 0% to 5% among various ethnic populations [63].
NQO1 AND VITAMIN E
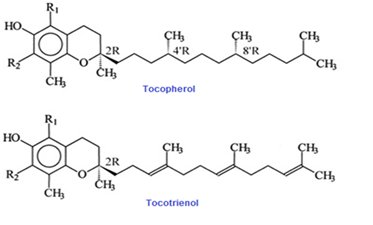
Vitamin E is an important group of compounds with multiple biological activities. It is broadly recognized to be the first line of antioxidant defense against lipid peroxidation preserving polyunsaturated fats in cellular membranes at the early stages of free radical attack through its free radical scavenging activity [64]. A total of eight dissimilar isoforms of vitamin E occur are identified to in nature that belongs into two classes; α, β, γ, δ tocopherols and α, β, γ, δ tocotrienols. The structure of these compounds contains a 6-chromanol ring and an isoprenoid side chain [65]. An isolated fraction of palm oil that contains primarily a mixture of α, γ, δ-tocotrienols and some α- tocopherols is referred to as “tocotrienol rich fraction” (TRF) [66]. Most of the tocopherols have saturated side-chains, whereas tocotrienols side chains possess double bonds at the 3', 7', and 11' positions (Figure 2).
Tocotrienols make up a substantial part of total vitamin E content in several food sources e.g. palm oil, rice bran and annatto (see table 1) [67]. The eight isoforms of vitamin E are absorbed into small intestinal epithelial cells via the lymphatic pathway and transported in association with chylomicrons into hepatic cells. Both tocopherols and tocotrienols undergo side-chain degradation initially by omega oxidation, followed by cycles of beta oxidation each decreasing the chain length by two carbons. The rate of metabolism of vitamin E is highest for tocotrienols and lowest for tocopherol, further suggesting on the tocotrienols' stronger biological activities [1].
There are claims that vitamin E, through its pronounced antioxidant activities, helps combat damaging free radicals. As such, it is expected to avert cells from any harmful by product of various metabolic pathways such as reactive oxygen species (ROS) [65]. All isomers of vitamin E possess antioxidant properties, however, tocotrienols have improved antioxidant activites in compare to tocopherols [68]. Further, trocrienols have other important functions, mainly in preserving a healthy cardiovascular system and a possible role in defense against cancer and other diseases [69]. The antioxidant activities of vitamin E has been linked with the induction of several phase II antioxidant enzymes involved in cancer [1]. These cytoprotective enzymes are essential for cell defense by enhancing the removal of free radicals and thus, play a protective role against cancer and other diseases [70]. NQO1 contains ARE (Antioxidant Response Element) sequence in the promoter region and therefore could hypothetically be regulated by nuclear factor (erythroid-derived 2)-like 2 (Nrf2) [4]. Nrf2 binds to ARE and regulate expression and induction of NQO1 gene [1]. Additionally, an association has been concluded between the antioxidant activity of tocotrienols with the redox sensitive Nrf2. Previous study has reported that tocotrienols were succeeded in controlling Nrf2-related phase II enzymes such as UDP-Glucuronyl transferase (UDP-GT), γ-glutamyl transferase (GGT) and glutathione S-transferase (GST) in breast cancer cells and mammary gland [71, 72, 73].
A previous study investigated the effects of α-tocopherol on phase II enzymes and its protective effects on acrolein-induced toxicity in human retinal pigment epithelial (RPE) cell line, ARPE-19 [74]. They found that pre-treatment with α-tocopherol activated the Nrf2/Keap1 pathway by enhancing the expression of Nrf2 and promoting its nuclear translocation. Consequently, the activities of several phase II detoxifying enzymes (such as NQO1) were demonstrated to be elevated [74]. Another study reported that the mRNA and protein expressions of NQO1 were induced in TRAMP-C1 cells treated with 30 mmol/L of γ-tocopherol-rich mixture of tocopherols compared with the control cells [75]. However, a different group of researchers found out that NQO1 expression was not affected by α-, γ- and δ-tocotrienols treatment in MCF-7 cells, but the expression of NQO2 was remarkably induced by tocotrienols [72]. Conversely, a nine-week treatment of vitamin E lacking diet to male rats resulted in enhanced NQO1 enzyme activities [76].
Data concerning to NQO1 gene expression and vitamin E have been limited. However, other phytochemicals and vitamins have been shown to impact NQO1 gene expression. Previous study had highlighted the role of NQO1 in preventing 17β- estradiol (E2)-induced breast cancer by antioxidants such as vitamin C (Vit C) or BHA or E2 metabolic inhibitor ANF in female ACI rats. The study showed that NQO1 activity was significantly attenuated in mammary tumors of E2-treated rats in compare to age-matched control mammary tissue. Whilst, the level of NQO1 enzyme activity was significantly elevated in mammary tissue of rats undergoing treatment with BHA, BHA + E2, Vit C or Vit C + E2 for 240 days, compared to control groups [77]. Increased NQO1 activity in kidneys, lungs and the mucosa of the upper small intestine was found in mice treated with vitamin E at a concentration of 7.5 g/kg for 14 days [51]. Other biochemical studies have elucidated that NQO1 activity is induced by a wide range of chemicals including polycyclic aromatic hydrocarbons, azo dyes and phenolic antioxidants [46].
Previous study has stated that treatment of animals with chemicals that generate mild oxidative stress in the liver does not cause any apparent liver damage, instead it shown to induce NQO1 and other antioxidant enzymes. Consequently, the increased level of antioxidant and detoxfying ability could aid in providing protection against the next exposure to oxidants and harmful chemicals and potential carcinogens [3]. This therapeutic strategy to diminish the risk of different types of cancer is referred to as chemoprevention, and has led to the development of diverse screening assays to characterize novel compounds that may able to suppress the carcinogenesis process in the liver or other organs [7]. One valuable screening method uses the induction of NQO1 enzyme activity in mouse Hepa-1c1c7 cell line as its end-point [78]. Through this assay, various chemicals (diphenols, quinones, dithiolethiones, hydroperoxides, peroxides, Michael reaction acceptors) were shown to induce NQO1 enzyme activity in mouse liver Hepa-1c1c7 cells [46]. Other many studies also used NQO1 induction Hepa- 1c1c7 cells in mouse liver to screen phytochemicals revealed that natural products can be used as potential anticancer agents. The findings of these studies manifested that chemicals obtained from natural products such as alkyl and alkenyl sulfides, carotenoids, brassinin derivatives, curcumin derivatives and flavonoids were able to elevate the level of NQO1 in mouse liver [3]. Induction of NQO1 is not unique to mouse Hepa-1c1c7 hepatoma cells but is also observed in non-transformed rat liver RL-34 cell lines [47].
The elevated level of NQO1 expression seen after TRF administration for a period of time can prevent liver damage due to excessive injury caused by the accumulation of poisonous toxins and their metabolites, in circumstances such as accidental ingestion aflatoxin-contaminated food, excessive alcohol intake, and chronic oral administration of over-the-counter drugs that could possibly cause injury to the liver such as NSAIDs. Previous studies had linked the antioxidant effects of tocotrienols with the ARE-Nrf2 system [71, 72, 73]. Therefore, it is postulated that the activation of ARE-Nrf2 signaling pathway by tocotrienols in TRF extract might be responsible for the up-regulation of NQO1 in mice liver.
CONCLUSIONS
NQO1, referred to as DT-diaphorase, is a homodimeric flavoenzyme that catalysis the beneficial two electron reduction of several quinones using flavin adenine dinucleotide (FAD) as a cofactor. NQO1 is mainly cytosolic enzyme exists as a homodimer and is biochemically characterized by its distinctive ability to use either NADH or NADPH as reducing cofactors and by its suppression by the anticoagulant dicumarol. It is frequently considered as a detoxifying enzyme due to of its ability to reduce reactive quinones and quinone imines to its forms of less active and less toxic hydroquinones. NQO1 is a considerably inducible enzyme that is regulated by the Nrf2/Keap1 system. Evidence for its implication in antioxidant activities provided by the appearances that induction or suppression of NQO1 levels are linked with decreased and increased exposures to oxidative stress, respectively. The NQO1 coding gene has two single nucleotide polymorphisms at position 609(C-T) and 465 (C-T) of the human cDNA. The presence of C465T causes reduction in enzyme activity, while the C609T causes profound loss of enzymatic activity due to protein instability.