My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Nutrición Hospitalaria
On-line version ISSN 1699-5198Print version ISSN 0212-1611
Nutr. Hosp. vol.28 n.2 Madrid Mar./Apr. 2013
https://dx.doi.org/10.3305/nh.2013.28.2.6424
ARTÍCULO ESPECIAL
Third Jesús Culebras Lecture - Molecular Biology and Clinical Nutrition; ¿where do we stand and where do we go?
Tercera lección Jesús Culebras - Biología molecular y nutrición clínica; ¿Dónde estamos y adónde vamos?
Ángel Gil
Departament of Biochemistry and Molecular Biology II. Institute of Nutrition and Food Technology "José Mataix". Biomedical Research Center. University of Granada. Armilla. Granada. Spain.
ABSTRACT
Nutrition plays a fundamental role in the maintenance of health and the treatment of disease, and serves as the crossroads for many disciplines. Biochemistry and Molecular Biology represents a key brand of science to ascertain the mechanism of action of nutrients and other food bioactive compounds in health and disease. The aim of the present Jesús M. Culebras lecture is to consider the future of the relationships between Molecular Biology and Clinical Nutrition and to discuss the use of molecular and genetic tools to study molecular responses to dietary factors and the metabolic consequences of food and to consider major challenges on human nutrition sciences in the 21st century. Particular emphasis is given to the identification and use of novel biomarkers in inflammatory diseases. Likewise, the importance of the human microbiome and how microorganisms can be safely utilized in the prevention and management of infectious and chronic diseases are discussed. Moreover, the key role of nutrigenetics, nutrigenomics and epigenetics in the new era of nutrition is considered. Nutrigenetics refers to the role of DNA sequence variation in the responses to nutrients, whereas nutrigenomics is the study of the role of nutrients in gene expression. Epigenetics is the study of mitotically heritable alterations in gene expression potential that are not caused by DNA sequence alterations. In the past decade, it has increasingly been recognized that dysregulation of epigenetic mechanisms may play an important role in human disease. Indeed, there is increasing interest in epigenetic mechanisms underlying phenotype modification modulated by nutrients. Further research in those areas should contribute to evaluate functionality of specific nutrients and bioactive compounds in Clinical Nutrition and allow personalized nutritional advice.
Key words: Biomarkers. Clinical nutrition. Epigenetics. Microbiota. Probiotics. Molecular biology. Nutrigenetics. Nutrigenomics.
RESUMEN
La Nutrición desempeña un papel fundamental en el mantenimiento de la salud y en el tratamiento de las enfermedades, sirviendo de encrucijada para muchas otras disciplinas. La Bioquímica y la Biología Molecular es una Ciencia clave para determinar los mecanismos de acción de los nutrientes y de otros compuestos bioactivos de los alimentos tanto en la salud como en la enfermedad. El objeto de esta 3a Lección Jesús M. Culebras es considerar el futuro de las relaciones de la Biología Molecular y de la Nutrición Clínica y comentar la utilización de las nuevas herramientas moleculares y genéticas en el estudio de las respuestas de los factores dietéticos y de las consecuencias metabólicas de la ingesta de alimentos, así como considerar los desafíos más importantes de la nutrición humana en el siglo XXI. En particular, se aborda la importancia de disponer de nuevos biomarcadores de interés nutricional en las enfermedades inflamatorias. Asimismo, se discute la importancia del microbioma humano y de cómo poder utilizar de forma segura los microorganismos en la prevención y el tratamiento de las enfermedades. Además, se considera la importancia clave de la nutrigenética, la nutrigenómica y la epigenética en la nueva era de la nutrición. La nutrigenética se refiere al papel de las variantes de secuencias del DNA en las respuestas de los individuos a los nutrientes, mientras que la nutrigenómica aborda el estudio de las modificaciones de expresión génica mediadas por los nutrientes. La epigenética es el estudio de las modificaciones en la expresión génica mediadas por las alteraciones del DNA heredables por mitosis. Durante la última década, se ha prestado importancia especial a la desregulación de los mecanismos epigenéticos causantes de enfermedad. Por tanto, existe un interés especial en conocer los mecanismos epigenéticos que modifican el fenotipo inducidos por los nutrientes.
Palabras clave: Biología molecular. Biomarcadores. Epigénetica. Microbiota. Nutrigenética. Nutrigenómica. Nutrición clínica. Probióticos.
Introduction
I am deeply honored to be the recipient of the Third Jesús M. Culebras Award. I met Prof. Culebras for the first time at the 3rd Congress of the Spanish Society of Parenteral and Enteral Nutrition (SENPE) held in Granada in May1986, organized by Dr. Antonio Perez de la Cruz. At that time I was involved in the identification of new biochemical biomarkers of nutritional status in low-birth weight infants1-3 and in the evaluation of the functional roles of dietary nucleotides, particularly on intestinal microbiota4 and on the lipoprotein and polyunsaturated fatty acid metabolisms in early life.5-7 Moreover, I was interested in the changes of plasma amino acids and polyunsaturated fatty acid (PUFA) profiles in severely trauma injured and infected children.8,9 In fact, one year later I joined the SENPE and I published my first two articles in Nutrición Hospitalaria, the journal created and directed by Prof. Culebras for now about 40 years, on the roles of dietary nucleotides in lipid metabolism in infants10 and on the effects on lipid metabolism of free-lipid parenteral nutrition in severely ill patients.11
At that Congress I also met Prof. Miquel A. Gassull and we discussed the possibility to starting to collaborate in the area of nutrition and major gastrointestinal diseases as in our lab we used modern tools in Biochemistry and they had patients with severe diseases, namely liver cirrhosis and inflammatory bowel disease. This was a large and long lasted collaboration of biochemists and gastroenterologists in Clinical Nutrition, which resulted in the identification of severe alterations of the polyunsaturated fatty acid metabolism in those diseases and novel approaches in the designing and utilization of modern enteral nutrition diets.12-15
I joined the Scientific and Educational Committee of the SENPE (CCE) by 1996 and started to work with Prof. Simon Schwartz and later with Dr. Mercè Planas and Dr. Julia Alvarez until 2010. During those 14 years I had the opportunity to interact, work and to become a friend of Prof. Jesús Culebras as he was, and he is, the Council member within the CEE representing the interests of Nutricion Hospitalaria, the official journal of SENPE. I always tried to collaborate and support him to get the dream of having a nutrition focused journal of recognized international interest, a feature he attained recently. Besides that, Prof. Culebras also supported me in the two editions of the Treatise of Nutrition (Tratado de Nutrición) and had a major role in my appointment as President of the Iberoamerican Council of Nutrition Journals in Montevideo, with the occasion of the 5th FELANPE Congress. Thank you very much to Jesús Culebras for giving me the opportunity to be his friend.
Nutrition plays a fundamental role in the maintenance of health and the treatment of disease, and serves as the crossroads for many disciplines. Human nutrition describes the processes whereby cellular organelles, cells, tissues organs, systems, and the body as a whole, obtain and use necessary substances obtained from foods (nutrients) to maintain structural and functional integrity and to growth and development. It also comprises studies on how nutrients interact with cell receptors and transporters and how are utilized at the molecular and cellular level and studies on gene-nutrition-environment interactions.16 Hence, Biochemistry and Molecular Biology represents a key brand of science to ascertain how nutrients and other food bioactive compounds are used by humans and what their influences at the molecular level are in order to be able to ascertain their mechanisms of action in health and disease.
During the second half of the 20th century Nutrition became a recognized multidisciplinary science that focused on the evaluation of the effects of nutrient deficiencies on organs, systems and human subjects. The major pathways by which nutrients are digested and metabolized were also identified. Likewise, a number of biochemical biomarkers associated with nutritional diseases were reported and described.17 Those biomarkers served as important tools to ascertain the specific biological effects of nutrients and their requirements in patients suffering many diseases and building up Clinical Nutrition as an important brunch of nutrition sciences.
The most important challenges of human nutrition in the 21st century is to know how nutrients and food bioactive compounds interact with different cell signaling cascades in different organs and systems and their specific effects on cell processes including gene activity regulation, metabolism, growth and apoptosis. Moreover, understanding how nutrients and other food components, as well as healthy lifestyle and environmental factors, are influencing the epigenetic processes and until what extent changes in the methylation pattern of DNA could be inherited represent an outstanding challenge for nutrition sciences. Furthermore, another major aim is to identify and ascertain how nutrients and food bioactive compounds are able to influence the human microbiome, and in turn how changes in the human microorganism ecology, particularly at the intestinal level, would have an influence in health an disease.
The main goal of the present lecture is to think about the future of relationships between Molecular Biology and Clinical Nutrition and to discuss the use of molecular and genetic tools to study molecular responses to dietary factors and the metabolic consequences of food and to consider major challenges on human nutrition sciences in the 21st century.
New biomarkers in nutrition: metabolomics can help
For the last decades numerous biomarkers have been identified and related to nutrient status in health and many diseases. In particular, many biomarkers are currently used in routine clinical nutrition to evaluate the nutritional status of populations, individual subjects and patients and to diagnose a number of pathologies and to assess the risk factors for certain pathologies.
Metabolomics is a metabolic-biology system approach focused on the metabolic responses understanding of living systems to physio-pathological stimuli by using multivariate statistical data on human body fluids obtained by different instrumental techniques. A metabolomic approach based on an analytical platform could be able to separate, detect, characterize and quantify a wide range of metabolites and its metabolic pathways. This instrumental approach has a good potential in the identification and detection of specific food intake and diseases biomarkers.18
In the present work it would be impossible to describe the new research concerning novel biomarkers of importance in nutrition. However, there some relevant findings that may serve to exemplify recent development in this area. Polyunsaturated fatty acids (PUFA) both of the omega-6 and omega-3 families have a profound influence on inflammation and regular intake of omega-3 fatty acids have a direct effect in the prevention of a number of inflammatory disorders including cardiovascular diseases, diabetes, rheumatoid arthritis and in the alleviation of symptoms for inflammatory bowel diseases as well as neurological disorders such as Alzheimer's disease and age-related macular degeneration.19,20 The resolution of inflammation is not a passive process, as believed earlier; instead, resolution is a biosynthetically active process, regulated by biochemical mediators and receptor-signaling pathways, and driven by specialized pro-resolving mediators (SPM). A number of findings by Serhan and his group, systematically investigated a number of SPM (biomarkers) derived from PUFA, including lipoxins, E-series resolvins, D-series resolvins, protectins/ neuroprotectins, and, most recently, maresins.21-23 These substances are mainly derived from eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA). The distinct properties of EPA and DHA to form primarily pro-resolving lipid mediators may explain their well-known beneficial health effects. These novel pathways may also explain some of the beneficial effects of aspirin, since they generate epimeric lipid mediators that are more metabolically stable and longer lasting.21
The human microbiome and the use of probiotics in Clinical Nutrition
A variety of microbial communities and their genes (the microbiome) exist throughout the human body, with fundamental roles in human health and disease.24-26 Diverse microbial communities from habitats within the human airways, skin, oral cavity, gut, and vagina can be found and microbiota may help to explain individual variability in health outcomes and be a source of new biomarkers for environmental exposures and of novel prognostic and diagnostic indicators.
The National Institutes of Health (NIH)-funded Human Microbiome Project Consortium has recently established a population-scale framework to develop metagenomic protocols, resulting in a broad range of quality-controlled resources and data including standardized methods for creating, processing and interpreting distinct types of high-throughput metagenomic data available to the scientific community.24,25 The resources from a population of 242 healthy adults sampled at 15 or 18 body sites up to three times, which have generated 5,177 microbial taxonomic profiles from 16S ribosomal RNA genes and over 3.5 terabases of metagenomic sequence have now been presented. In parallel, approximately 800 reference strains isolated from the human body have been sequenced. Collectively the data represent a treasure trove that can be mined toidentify new organisms, gene functions, and metabolic and regulatory networks, as well as correlations between microbial community structure and health and disease. Among other future benefits, this resource should contribute to promote the development of novel prophylactic strategies such as the application of prebiotics and probiotics to foster human health.24
Microbes in the human gut undergo selective pressure from the host as well as from microbial competitors. This typically leads to a homeostasis of the ecosystem in which some species occur in high and many in low abundance, with some low abundance species, performing specialized functions beneficial to the host.27 Over the past few years, the application of next-generation sequencing approaches (metagenomics) to the study of human-associated microorganisms has shown that the composition of the microbiota within the guts of different individuals is distinct. This distinctiveness is possible because of the marked variability that is evident at the species and strain levels within the microbiota. By contrast, variability at the phylum level is not individual specific. Indeed, more than 90% of gut bacteria are members of only two phyla, Bacteroidetes and Firmicutes, and the relative proportions of these two major divisions exhibit a continuous gradient within the human population, with some individuals having a predominance of the former, others having a predominance of the latter, but the majority having similar proportions of each.24-26
The very recent studies of the human microbiome) have revealed that even healthy individuals differ remarkably in the microbes that occupy habitats such as the gut, skin and vagina.24,25 Much of this diversity remains unexplained, although diet, environment, host genetics and early microbial exposure have all been implicated. Recently, it has been reported that the diversity and abundance of each habitat's signature microbes to vary widely even among healthy subjects, with strong niche specialization both within and among individuals.26,28 These results delineate the range of structural and functional configurations normal in the microbial communities of a healthy population, enabling future characterization of the epidemiology, ecology and translational applications of the human microbiome.
In an apparent contrast to the phylum distributions, it has been suggested that variation at the species level is discontinuous, with three clusters, or enterotypes, that vary in proportional composition.27 This proposal was based on multidimensional cluster analysis and principal component analysis of faecal metagenomes from 39 samples involving six nationalities and including 22 newly sequenced European faecal samples. The proposed enterotypes were identified by their enrichment in Bacteroides spp. (enterotype 1), Prevotella spp. (enterotype 2) and Ruminococcus spp. (enterotype 3), and were unrelated to nationality or host characteristics such as body mass index, age or gender. It was subsequently suggested that 'faecotypes' might have been a more accurate descriptor than enterotypes, as microbial composition and abundance varies along the gastrointestinal tract.29
Shortly after the enterotype concept was mooted,27 Wu et al.30 showed that in subjects aged 2-50 years old, two enterotypes were associated with diet: long-term diets enriched for protein and animal fat were associated with the Bacteroides enterotype, whereas diets enriched for carbohydrate were associated with the Prevotella enterotype.
Although the enterotype distinctions appear to be less clear than was first thought, and regardless of whether we are dealing with gradients or clusters, traits or states, the concept of linking patterns of microbial composition with function is both biologically plausible and clinically relevant.28 We need to move from retrospective correlative analysis to prospective studies linking microbiota variation with clinically relevant outcomes. Categorizing the microbiota into discrete groups would be attractive if these groups overlapped with, for example, responders or non-responders to particular therapeutics (reflecting the role of the gut microbiota as a bioreactor capable of drug modification)or with rates of progression in immune-mediated disease. The struggle to clarify microbial complexity within the gut and to link it to clinical traits should continue. It will require longitudinal studies of different populations with different lifestyles and dietary patterns so that clustering, transitions and intermediates can be identified.
Currently, there is an increasing interest in and demand for probiotics, after a long history of safe use in fermented dairy products and an increased recognition of the beneficial effects of probiotics to human gut health.31 According to the Food and Agriculture Organisation of the United Nations and the World Health Organisation,32 probiotics are "live microorganisms which, when administered in adequate amounts, confer a health benefit on the host." In particular, strains belonging to Bifidobacterium and Lactobacillus, the predominant and subdominant groups of the gastrointestinal microbiota, respectively,33 are the most widely used probiotic bacteria and are included in many functional foods and dietary supplements.34-36 The yeast Saccharomyces boulardii has also been shown to have health benefits.37
For probiotics to be successful, they must possess certain characteristics. The criteria for the selection of probiotics include tolerance to gastrointestinal conditions (gastric acid and bile), ability to adhere to the gastrointestinal mucosa and competitive exclusion of pathogens.38-40
The results of evidence-based analyses from human studies and animal models have shown the potential clinical effectiveness of probiotics on many diseases.41 In fact, probiotics have been reported to suppress diarrhoea,42 alleviate lactose intolerance43 and postoperative complications,14 exhibit antimicrobial45 and anticolorectal cancer activities,46,47 reduce irritable bowel symptoms48 and prevent inflammatory bowel disease.49 However, generalisations concerning the potential health benefits of probiotics should be not made because probiotic effects tend to be strain-specific; thus, the health benefit attributed to one strain is not necessarily applicable to another strain, even within one species.50
Probiotics have been shown to promote a variety of biological effects in a number of physiological conditions and pathologies, including allergy, intestinal and liver diseases, urinary and upper respiratory infections, AIDS and metabolic diseases. These effects are strain-specific and are primarily mediated through changes in the faecal microbiota and immune modulation. RCTs concerning the appropriate clinical evaluation of probiotics with an adequate and statistically sufficient number of subjects related to main outcome variables should be performed in a variety of diseases. In addition, multicentre and replicate studies are necessary to evaluate the actual role of probiotics in the amelioration of symptoms for many diseases. The number of studies concerning the mechanism of probiotics in cell and animal models is scarce. Apparently, many probiotics are able to modulate both the innate and adaptive immune responses; however, the molecular basis of these effects remains unknown.51
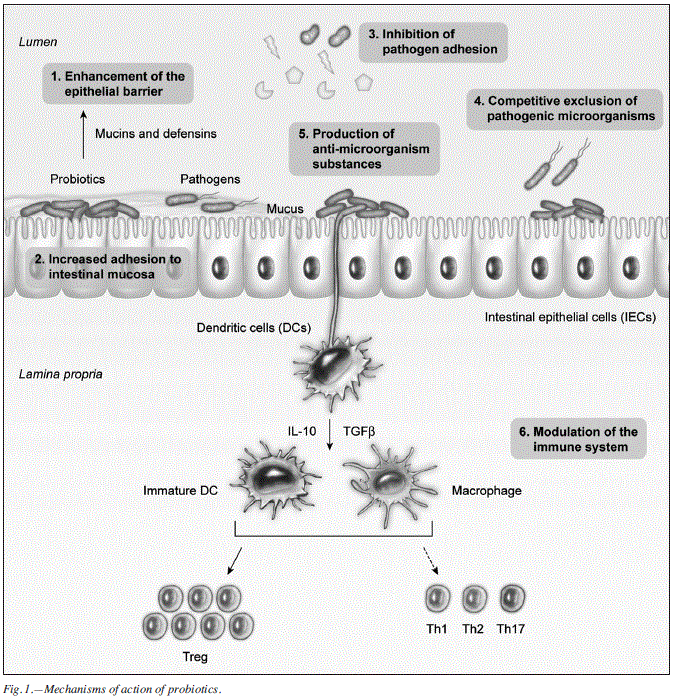
The mechanisms underlying the beneficial effects of probiotics are largely unknown but are likely to be multifactorial, including modificationof the gut microbiota, competitive adherence to the mucosa and epithelium, strengthening of the gut epithelial barrier and modulation of the immune system to convey an advantage to the host (fig. 1).52 Accumulating evidence demonstrates that probiotics communicate with the host by pattern recognition receptors, such as toll-like receptors and nucleotide binding oligomerization domain-containing protein-like receptors, which modulate key signaling pathways, such as nuclear factor -κB and mitogen-activated protein kinase, to enhance or suppress activation and influence downstream pathways. This recognition is crucial for eliciting measured antimicrobial responses with minimal inflammation damage. A clear understanding of these mechanisms will allow for appropriate probiotic strain selection for specific applications and may uncover novel probiotic functions.52
Nutrigenomics and nutrigenetics: are we ready for personalised nutrition?
The efficacy by which dietary interventions influence risk markers of multi-factorial diseases is mainly determined by taking population-based approaches. However, there exists considerable inter-individual variation in response to dietary interventions, and some interventions may benefit certain individuals or population subgroups more than others.53
The execution of the Human Genome Project has brought forth a wealth of information about the structure of the genome, which can now be used to study how the interplay between our genes and factors from the environment such as nutrition relate to a state of health or disease. To enable such studies, novel technologies have been designed in particular to monitor the activity of multiple genes simultaneously at the level of the RNA by transcriptomics, or the level of the proteins by proteomics (fig. 2). In addition, genome information has boosted approaches to study the role of genetic variation to explain individual differences in responses to nutrition, underlying in part the susceptibility for nutrition-related disorders.
Nutrigenetics refers to the role of DNA sequence variation in the responses to nutrients, whereas nutrigenomics is the study of the role of nutrients in gene expression (fig. 3). This research is predicated on the assumption that there are individual differences in responsiveness to acute or repeated exposures to a given nutrient or combination of nutrients. Throughout human history, diet has affected the expression of genes, resulting in phenotypes that are able to successfully respond to environmental challenges and that allow better exploitation of food resources. These adaptations have been key to human growth and development. Technological advances have made it possible to investigate not only specific genes but also to explore in unbiased designs the whole genome-wide complement of DNA sequence variants or transcriptome. These advances provide an opportunity to establish the foundation for incorporating biological individuality into dietary recommendations, with significant therapeutic potential i.e. personalized nutrition.53-54
The influence of nutrition on genome activity is studied almost always in a comparative manner either by a direct or an indirect approach. The direct approach involves changes in the nutrients presented to a model system followed by monitoring the changes in gene expression. The indirect approach involves the study of nutrition-related traits and disorders such as obesity, type II diabetes and cardiovascular disorders. In those studies, gene expression is compared between subjects with and without the disorder and, from the differences, scientists hope to deduce the relevant molecular pathways leading from health to disease under the influence of diet and lifestyle.55 The results of those studies should lead to new targets for pharmacological or dietary intervention and to novel functional foods.56
One of the possibilities of nutrigenomic technologies to further the concept of personalised nutrition, as well as the process to take personalised nutrition to the marketplace. The modulation of an individual's response is influenced by both genetic and environmental factors. Many nutrigenetics studies have attempted to explain variability in responses based on a single or a few genotypes so that a genotype may be used to define personalised dietary advice. It has, however, proven very challenging to define an individual's responsiveness to complex diets based on common genetic variations. In addition, there is a limited understanding of what constitutes an optimal response because we lack key health biomarkers and signatures. In conclusion, advances in nutrigenomics will undoubtedly further the understanding of the complex interplay between genotype, phenotype and environment, which are required to enhance the development of personalised nutrition in the future. At the same time, however, issues relating to consumer acceptance, privacy protection as well as marketing and distribution of personalised products need to be addressed before personalised nutrition can become commercially viable.53
Nutritional advice has mainly focused on population-level recommendations. Recent developments in nutrition, communication, and marketing sciences have enabled potential deviations from this dominant business model in the direction of personalisation of nutrition advice. Such personalisation efforts can take on many forms, but these have in common that they can only be effective if they are supported by a viable business model. Future research should explore the consumer responses to the diversity of "archetypical" business models for personalised nutrition advice as a source of market information on which the delivery of nutrigenomics-based personalised nutrition advice may further build.57
The relationships between diet and nutrigenomic-metabolomic profiles, as well as between these profiles and health, are being elucidated, and this will dramatically alter clinical practice in nutrition. In fact, nutrigenomics and metabolomics provide methodology that allows clinicians to view a broader footprint of what is going on in metabolism than they can get using current clinical chemistry panels. This could greatly refine the practice of clinical nutrition. When a nutrition clinical trial is conducted, nutrigenomic methods can help investigators to understand why a subgroup of study subjects responded to treatment, while others did not. This could reduce the "noise" that often clouds such clinical studies.58
Epigenetics is the study of mitotically heritable alterations in gene expression potential that are not caused by DNA sequence alterations.58 By stably regulating gene expression potential in differentiated tissues, epigenetic mechanisms such as DNA methylation play a critical role in mammalian development (fig. 4). In the past decade, it has increasingly been recognized that dysregulation of epigenetic mechanisms may play an important role in human disease.60 Indeed, there is increasing interest in epigenetic mechanisms underlying phenotype modification modulated by nutrients (fig. 5). Further research in this area should contribute to evaluate functionality of specific nutrients and bioactive compounds in Clinical Nutrition.
Conclusions
New biomarkers useful in the diagnosis and follow-up of chronic diseases and in the evaluation of nutritional treatments are expanding due to the use of metabolomics opening a new era in Clinical Nutrition
The identification of human microbiome and its functions as well as the ascertaining of the mechanisms underlying the effects of probiotics is opening new perspectives to the use of microorganims in the prevention and treatment of chronic diseases.
The new "omic" sciences i.e. transcriptomics, proteomics and metabolomics are allowing to determine the interactions between nutrients and other bioactive food components and genes. This would contribute to a better treatment of diseases and to a personalized nutrition.
The incidence of chronic diseases in the adult is related to epigenetic changes that can occur even in early life. The prevention of those diseases through appropriate interactions between diet, the environment and the host constitutes one of the biggest challenges of nutrition in the 21st Century.
References
1. Faus MJ, Gil A, Robles R, Sánchez-Pozo A, Pita ML, Sánchez-Medina F. Changes in serum albumin, transferrin and amino acid indices during the first month of life in small-for-date infants. Ann Nutr Metab 1984; 28: 70-76. [ Links ]
2. Robles R, Gil A, Faus MJ, Periago JL, Sánchez-Pozo A, Pita ML, Sánchez-Medina F. Serum and urine amino acid patterns during the first month of life in small-for-date infants. Biol Neon 1984; 45: 209-217. [ Links ]
3. Gil A, Faus MJ, Robles R, Pita ML, Sánchez-Pozo A, Linares MD, Sánchez-Medina F. Urinary 3-methylhistidine derivative as indicator of nutrients intake in low-birth-weight infants. Horm Met Res 1984; 16: 667-670. [ Links ]
4. Gil A, Corral E, Martínez A, Molina JA. Effects of the addition of nucleotides to an adapted milk formula on the microbial pattern of faeces in at term newborn infants. J Clin Nutr Gastroenterol 1986; 1: 127-132. [ Links ]
5. Gil A, Pita ML, Martínez A, Molina JA, Sánchez-Medina F. Effect of dietary nucleotides on the plasma fatty acids in at-term neonates. Human Nutr: Clin Nutr (currently Eur J clin Nutr) 1986; 40C: 185-195. [ Links ]
6. Sánchez-Pozo A, Pita ML, Martínez A, Molina JA, Sánchez-Medina F, Gil A. Effects of dietary nucleotides upon lipoprotein pattern of newborn infants. Nutr Res 1986; 6: 763-771. [ Links ]
7. De-Lucchi C, Pita ML, Faus MJ, Molina JA, Uauy R, Gil A. Effects of dietary nucleotides on the fatty acid composition of erythrocyte membrane lipids in term infants. J Ped Gastroenterol Nutr 1987; 6: 568-574. [ Links ]
8. Maldonado J, Pita ML, Narbona E, Gil A, Molina JA. Changes in the serum fatty acid patterns of injured and infected children receiving fat-free parenteral nutrition. J Clin Nutr Gastroenterol 1987; 2: 105-112. [ Links ]
9. Maldonado J, Gil A, Faus MJ, Pita ML, Molina JA. Serum fatty acids and amino acids. Are they markers of the severity and outcome in trauma children? J Clin Nutr Gastroenterol 1987; 2: 74-80. [ Links ]
10. Gil A, Medina A, Pita ML, De-Lucchi C, Martínez-Valverde A. Influencia de los nucleótidos de la dieta en el metabolismo de los ácidos grasos poliinsaturados en recién nacidos pretérmino. Nutr Hosp 1987; 4: 164-168. [ Links ]
11. Gil A, Maldonado J, Pita ML, De-Lucchi C, Molina JA. Ácidos grasos séricos en niños con politraumatismo: Efectos de la nutrición parenteral exenta de lípidos. Nutr Hosp 1987; 4: 11-16. [ Links ]
12. Cabré E, Periago JL, Abad-Lacruz A, Gil A, González-Huix F, Sánchez-Medina F, Gassull MA. Polyunsaturated fatty acid deficiency in liver cirrhosis: Its relation to associated protein-energy malnutrition (Preliminary report). Am J Gastroenterol 1988; 83 (7): 712-717. [ Links ]
13. Abad-Lacruz A, Fernández-Bañares F, Cabré E, Gil A, Esteve M, González-Huix F, Xiol X, Gassull MA. The effect of total enteral tube feeding on the vitamin status of malnourished patients with inflammatory bowel disease. Int J Vit Nutr Res 1988; 58: 428-435. [ Links ]
14. Cabré E, Periago JL, Abad-Lacruz MD, González-Huix F, González J, Esteve M, Fernández-Bañares F, Planas R, Gil A, Sánchez-Medina F, Gassull MA. Plasma fatty acid profile in advanced cirrhosis: Unsaturation deficit of lipid fractions. Am J Gastroenterol 1990; 85: 1597-1604. [ Links ]
15. Fernández-Bañarés F, Mingorance MD, Esteve M, Cabré E, Lachica M, Abad A, Gil A, Humbert P, Boix J, Gassull MA. Serum zinc, copper, and selenium levels in inflammatory bowel disease. Effect of total enteral nutrition on trace elements status Am J Gastroenterol 1990; 85: 1584-1589. [ Links ]
16. Gil A. Tratado de Nutrición. 2a Ed Editorial Panamericana, Madrid, 2010 (volumes 1 to 4). [ Links ]
17. Raqib R, Cravioto A. Nutrition, immunology, and genetics: future perspectives. Nutr Rev 2009; 67(Suppl. 2): S227-S236. [ Links ]
18. Vernocchi P, Vannini L, Gottardi D, Del Chierico F, Serrazanetti DI, Ndagijimana M, Guerzoni ME. Integration of datasets from different analytical techniques to assess the impact of nutrition on human metabolome. Front Cell Infect Microbiol 2012; 2: 156. [ Links ]
19. Gil A, Serra-Majem L, Calder PC, Uauy R. Systematic reviews of the role of omega-3 fatty acids in the prevention and treatment of disease. Br J Nutr 2012; 107 (Suppl. 2): S1-2. [ Links ]
20. Rangel-Huerta OD, Aguilera CM, Mesa MD, Gil A. Omega-3 long-chain polyunsaturated fatty acids supplementation on inflammatory biomarkers: a systematic review of randomised clinical trials. Br J Nutr 2012; 107 (Suppl. 2): S159-70. [ Links ]
21. Serhan CN. The resolution of inflammation: the devil in the flask and in the details. FASEB J 2011; 25: 1441-1448. [ Links ]
22. Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chem Rev 2011; 111: 5922-5943. [ Links ]
23. Buckley CD, Gilroy DW, Serhan CN, Stockinger B, Tak PP. The resolution of inflammation. Nat Rev Immunol 2013; 13: 59-66. [ Links ]
24. Methé BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower Cand The Human Microbiome Project Consortium. Aframework for human microbiome research. Nature 2012; 486: 215-221. [ Links ]
25. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT and The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012; 86: 207-214. [ Links ]
26. Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012; 488 (7410): 178-84. [ Links ]
27. Arumugam M, Raes J, Pelletier E, Paslier DL, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature 2011; 73: 174-180. [ Links ]
28. O'Toole's PW. Categorization of the gut microbiota: enterotypes or gradients? Nature Rev Microbiol 2012; 10: 591-592. [ Links ]
29. Siezen RJ, Kleerebezem M. The human gut microbiome: are we our enterotypes? Microb Biotechnol 2011; 4: 550-553. [ Links ]
30. Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011; 334: 105-108. [ Links ]
31. Liong MT. Probiotics: Biology, Genetics and Health aspects. Microbiol Monographs 2011. [ Links ]
32. FAO/WHO. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. 2011 www.fao.org [ Links ]
33. Guarner F, Malagelada JR. Gut flora in health and disease. Lancet 2003; 361: 512-519. [ Links ]
34. Gourbeyre P, Denery S, Bodinier M. Probiotics, prebiotics, and synbiotics: impact on the gut immune system and allergic reactions. J Leukoc Biol 2011; 89: 685-695. [ Links ]
35. Macpherson AJ, Harris NL. Interactions between comensal intestinal bacteria and the immune system. Nat Rev Immunol 2004; 4: 478-485. [ Links ]
36. Frick JS, Schenk K, Quitadamo M, et al. Lactobacillus fermentum attenuates the proinflammatory effect of Yersinia enterocolitica on human epithelial cells. Inflamm Bowel Dis 2007;13:83-90. [ Links ]
37. McFarland. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol 2006; 101: 812-822. [ Links ]
38. Collins JK, Thornton G, Sullivan GO. Selection of probiotic strains for human application. Int Dairy J 1998; 8: 487-490. [ Links ]
39. Ouwehand AC, Salminen S, Isolauri E. Probiotics: an overview of beneficial effects. Antonie van Leeuwenhoek 2002; 82: 279-289. [ Links ]
40. Collado MC, Gueimonde M, Salminen S. Probiotics in Adhesion of Pathogens: Mechanisms of Action. Bioactive Foods in Promoting Health 2010; 23: 353-370. [ Links ]
41. Yan F, Polk DB. Probiotics and immune health. Curr Opin Gastroenterol 2011; 27: 496-501. [ Links ]
42. Lye HS, Kuan CY, Ewe JA, et al. The improvement of hypertension by probiotics: effects on cholesterol, diabetes, renin, and phytoestrogens. Int J Mol Sci 2009; 10: 3755-3775. [ Links ]
43. Pelletier X, Laure-Boussuge S, Donazzolo Y. Hydrogen excretion upon ingestion of dairy products in lactose-intolerant male subjects: importance of the live flora. Eur J Clin Nutr 2011; 55: 509-512. [ Links ]
44. Woodard GA, Encarnacion B, Downey JR, et al. Probiotics improve outcomes after Roux-en-Y gastric bypass surgery: a prospective randomized trial. J Gastrointest Surg 2009; 13:1198-1204. [ Links ]
45. Karska-Wysocki B, Bazo M, Smoragiewicz W. Antibacterial activity of Lactobacillus acidophilus and Lactobacillus casei against methicillin-resistant Staphylococcus aureus (MRSA). Microbiol Res 2010; 165: 674-686. [ Links ]
46. Liong MT. Safety of probiotics: translocation and infection. Nutr Rev 2008; 66: 192-202. [ Links ]
47. Rafter J, Bennett M, Caderni G, et al. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am J Clin Nutr 2007; 85: 488-496. [ Links ]
48. Moayyedi P, Ford AC, Talley NJ, et al. The efficacy of probiotics in the treatment of irritable bowel syndrome: a systematic review. Gut 2010; 59: 325-332. [ Links ]
49. Golowczyc MA, Mobili P, Garrote GL, et al. Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar enteritidis. Int J Food Microbiol 2007; 118: 264-273. [ Links ]
50. Williams NT. Probiotics. Am J Health-System Pharm 2010; 67: 449-458. [ Links ]
51. Fontana L, Bermudez-Brito M, Plaza-Diaz J, Muñoz-Quezada S, Gil A. Sources, isolation, characterisation and evaluation of probiotics. Br J Nutr 2013; 109 (Suppl. 2): S35-S50. [ Links ]
52. Bermudez-Brito M, Plaza-Díaz J, Muñoz-Quezada S, Gómez-Llorente C, Gil A. Probiotic mechanisms of action. Ann Nutr Metab 2012; 61: 160-174. [ Links ]
53. de Roos B. Personalised nutrition: ready for practice? Proc Nutr Soc 2012; 12: 1-5. [ Links ]
54. Bouchard C, Ordovas JM. Fundamentals of nutrigenetics and nutrigenomics. Prog Mol Biol Transl Sci 2012; 108: 1-15. [ Links ]
55. Olza J, Gil-Campos M, Leis R, Rupérez AI, Tojo R, Cañete R, Gil A, Aguilera CM. A gene variant of 11 beta-hydroxysteroid dehydrogenase type 1 is associated with obesity in children. Int J Obes (Lond) 2012; 36 (12): 1558-63. [ Links ]
56. Mariman ECM. Nutrigenomics and nutrigenetics: the 'omics' revolution in nutritional science. Biotechnol Appl Biochem 2006; 44: 119-128. [ Links ]
57. Ronteltap A, van Trijp H, Berezowska A, Goossens J. Nutrigenomics-based personalised nutritional advice: in search of a business model? Genes Nutr 2012 (Epub ahead of print). [ Links ]
58. Zeisel SH, Waterland RA, Ordovás JM, Muoio DM, Jia W, Fodor A. Highlights of the 2012 Research Workshop: Using Nutrigenomics and Metabolomics in Clinical Nutrition Research. JPEN J Parenter Enteral Nutr 2012 (Epub ahead of print). [ Links ]
59. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003; 33 (Suppl.): 245-254. [ Links ]
60. Gluckman PD, Hanson MA, Buklijas T, Low FM, Beedle AS. Epigenetic mechanisms that underpin metabolic and cardiovascular diseases. Nat Rev Endocrinol 2009; 5: 401-408. [ Links ]
![]() Correspondence:
Correspondence:
Ángel Gil.
Instituto de Nutrición y Tecnología de los Alimentos "José Mataix" (INyTA).
Centro de Investigación Biomédica (CIBM).
Universidad de Granada.
Avenida del Conocimiento s/n.
18100 Armilla. Granada. España.
E-mail: agil@ugr.es
Recibido: 12-XI-2012.
Aceptado: 26-XI-2012.