Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Revista Española de Enfermedades Digestivas
versión impresa ISSN 1130-0108
Rev. esp. enferm. dig. vol.102 no.1 Madrid ene. 2010
Vascular endothelial growth factor gene polymorphisms in patients with colorectal cancer
Polimorfismos del gen del factor de crecimiento vascular endotelial en pacientes con cáncer colorrectal
M. Vidaurreta1, R. Sánchez-Muñoz2, S. Veganzones1, S. Rafael1, M. Gutiérrez1, V. de-la-Orden1, C. Fernández3, M. Arroyo1, F. J. Cerdán2 and M. Maestro1
Departments of 1Clinical Analysis, 2General Surgery and Digestive Diseases, and 3Research Unit. Hospital Clínico San Carlos. Madrid, Spain
ABSTRACT
Background: angiogenesis plays an important role in tumor progression. The vascular endothelial growth factor (VEGF) is an important regulator of angiogenesis. In the present study we evaluated single nucleotide polymorphisms (SNPs) -2578C > A, -1154G > A, and +936C > T in the VEGF gene, and their prognostic value for patients operated on for colorectal cancer (CRC).
Patients and method: VEGF polymorphisms have been analyzed in 177 patients who had undergone surgical resection at Hospital Clínico San Carlos. The analysis of these polymorphisms was performed with specific probes for each nucleotide in a multiplex reaction using real-time PCR.
Results: we only found a statistically significant relationship for one of these three polymorphisms, +936C > T, with gender and tumor location; 10.7% of patients heterozygotes for this SNP had tumors located in proximal colon, 35.2% in distal segment and 54.1% in rectum (p = 0.03). Patients with the +936T/T genotype had 100% overall survival (OS).
Conclusion: patients with a +936T/T genotype showed increased survival, therefore the +936C > T SNP could be a useful marker in the follow-up and clinical management of patients with colorectal cancer.
Key words: VEGF polymorphisms. Colorectal cancer.
RESUMEN
Introducción: la angiogénesis juega un papel importante en la progresión de los tumores. El factor de crecimiento endotelial vascular (VEGF) es un importante regulador de la angiogénesis. En este trabajo se han analizado los polimorfismos de único nucleó-tido (SNP) -2578C > A, -1154G > A y +936C > T del gen VEGF en pacientes intervenidos de carcinoma colorrectal, así como su posible implicación pronóstica.
Pacientes y método: el estudio de estos SNP se ha realizado en 177 pacientes intervenidos quirúrgicamente de carcinoma colorrectal (CCR) en el Hospital Clínico San Carlos. El análisis de los polimorfismos se realizó con sondas específicas para cada nucleótido y se determinó mediante una reacción multiplex mediante real time PCR.
Resultados: de los 3 polimorfismos estudiados sólo encontramos relación estadísticamente significativa del SNP +936C > T con el sexo y la localización. El 10,7% de los pacientes heterocigotos para este SNP tenían como localización el tumor en colon proximal, el 35,2% en colon distal y el resto en recto (p = 0,03). La supervivencia global (SG) de los pacientes con el genotipo +936T/T fue del 100%.
Conclusión: los pacientes con el genotipo +936T/T presentan mayor supervivencia y el polimorfismo +936C > T podría ser una herramienta de ayuda en el seguimiento y terapéutica de este grupo de pacientes.
Palabras clave: VEGF polimorfismos. Cáncer colorrectal.
Introduction
Angiogenesis is a prerequisite for the development, growth and progression of solid tumors (1). The vascular endothelial growth factor (VEGF) is one of the most potent endothelial cell mitogens and plays and important role in angiogenesis (2). Different clinical studies have shown that high levels of VEGF expression and increased microvessel density in tumors were associated with advanced stage disease and worse prognosis for various types of tumors, including colorectal cancer (CRC) (3-5).
The VEGF gene is located on chromosome 6p21.3 and consists of eight exons that exhibit alternative splicing to form a family of proteins (6,7). Several single nucleotide polymorphisms (SNPs) in VEGF have been reported to affect the expression of this gene. These SNPs have been associated with variations in VEGF protein production and reported to be involved in the development of several tumors (8,9). Some of these SNPs [-2578C > A, -1154G > A, and -634G > C in the promoter region and +936C > T in the 3'-untranslated region] were found to be associated with variations in VEGF protein synthesis (7,10). The -2578C/A, -1154G/G y -634G/C genotypes appear to be associated with higher VEGF expression (11,12), whereas the +936T allele is correlated to lower VEGF expression (10).
The aim of this study was to analyze three VEGF gene polymorphisms: -2578C/A, -1154G/A and +936C/T. We also evaluated the relationship of these SNPs with clinicopathologic variables, and its possible implication for prognosis in a population of patients operated on for CRC.
Patients and methods
Study population
The study cohort was comprised of 177 patients undergoing surgery consecutively for primary CRC at Hospital Clínico San Carlos, Madrid (Spain) between March 1995 and January 2000. This was a prospective cohort study. All patients were operated on by the same surgeon, who performed radical oncological surgery based on the location of the tumor. The surgery was defined as curative when there was no evidence of macroscopic residual tumor, no distant metastases were found, and in the pathological analysis, no invaded margins were detected. Using this criterion, the surgeon performed a curative resection in 145 patients (81.9%) and resected the primary tumor in 32 patients (18.1%) as palliative treatment. Patients with metacronic carcinoma, familial polyposis, hereditary nonpolyposis colon cancer (HNPCC), and inflammatory bowel disease were excluded from the study. None of the patients had received neoadjuvant treatment. An informed consent was obtained from each patient, and the project was approved by the clinical research and ethics committee of this hospital. Follow-up was performed according to the protocol designed by the authors (13). The variables considered in the study were sex, age, Dukes' stage, tumor site, differentiation grade, histological type, adjuvant chemotherapy, and VEGF gene SNPs. Tumors were staged according to Duke's classification. Proximal tumors were defined as occurring from the cecum through to the transverse colon; tumors in the splenic flexure, descending and sigmoid colon were defined as distal. Stage B and C patients received adjuvant treatment with 5-fluorouracil (5-FU) and leucovorin (75% of patients included). For stage D patients different protocols were applied according to Oncology Service criteria.
Tumoral and non-tumoral tissue samples were obtained during the surgical procedure and immediately immersed in liquid nitrogen for storage in a freezer at -80 oC. Specimens were then independently examined by two pathologists, who confirmed the samples had over 80% tumor cells.
For genetic analyses, genomic DNA was isolated from tumoral and non-tumoral tissue samples using the DNasy® Blood & Tissue Kit (Qiagen) according to the manufacturer's instructions.
Genotyping of VEGF gene polymorphisms
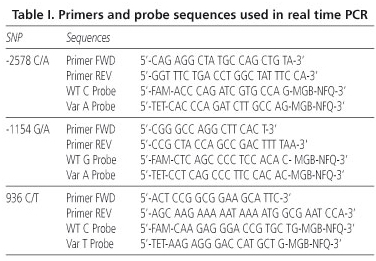
In this study, -2578C > A, -1154G > A and +936C > T polymorphisms were determined by TaqMan® MGB probes based on polymerase chain reaction. Sequences of forward and reverse primers and probes for all three polymorphisms are listed in table I (14). The different genotypes were evaluated by quantitative real-time PCR in a multiplex reaction using a Smart Cycler (CEPHEID, Sunnyvale, USA).
Statistical analysis
Qualitative variables were provided with their corresponding frequency distributions. Quantitative variables were expressed as mean, standard deviation (SD) and range. All variables were qualitative except age, overall survival (OS), and disease-free survival (DFS). Age was recorded into two groups according to median age (71 years). Associations between qualitative variables were evaluated using the c2 test or Fisher's exact test when 25% of expected frequencies fell below 5. Overall survival (OS) was estimated using the Kaplan-Meier method and compared between groups using Breslow's exact test. An event in OS was defined as death occurring as a consequence of the tumor, censuring live patients and those dying of another cause. OS was calculated as the time elapsed from the date of surgery to death or last follow-up. Data were fitted into Cox's proportional risk regression model. The hazard ratio (HR) was given with a 95% (95% CI) confidence interval. The variables included according to biological criteria were: sex, age, Duke's stage, tumor site, differentiation grade, histological type, adjuvant chemotherapy, and SNPs for the VEGF gene. In each comparison the null hypothesis was rejected when the type I error was less than 0.05. All statistical tests were performed using the SSPS v.o 11.5 software (SPSS.Inc Chicago, ILL III).
Results
The study cohort comprised 177 patients, 52.1% of whom were male and 46.3% female. Mean patient age was 71 years (median 71 years; range 35-94 years). Clinicopathological variables are shown in table II. With respect to location, 25.7% of tumors were located in the proximal colon, 29.5% in the distal colon, and 44.8% in the rectum. Most of the tumors analyzed were non-mucinous adenocarcinomas, and only 8.7% were mucinous. In 21 patients differentiation grade could not be established.
The distribution of genotypes for each VEGF polymorphism analyzed and their allelic frequencies are shown in table III. In one patient -1154G > A SNP could not be determined; 27.7% of patients showed A/A genotype in -2578C > A SNP, 22.2% showed genotype A/A in -1154G > A SNP, and 1.1% showed a T/T genotype in 936C > T; 36.7% of patients were heterozygotes for the -2578C > A SNP, 33.0% for the -1154G > A SNP, and 25.4% for the +936C > T SNP.
In the analysis of clinicophatological variables and VEGF SNPs, only the T/T genotype in the +936C > T SNP was related to sex and tumor location. The two patients who showed a T/T genotype were women. The frequency of the T/T genotype in women was higher than in men (2.4 vs. 0%), and the frequency of the C/T genotype was significantly lower (18.3 vs. 31.6%) (p = 0.03). In 10.7% of patients who were heterozygotes for the +936C > T SNP had tumors located in the proximal colon, 35.2% in the distal colon, and 27.6% in the rectum (p = 0.03). No relation was found for other clinicopathological variables (Table II).
In localized tumors (stages A and B) we found a higher percentage of T/T genotypes in the +936C > T SNP, although no significant relation with stage was found for this SNP (p = 0.08).
The presence of a T allele (C/T and T/T genotypes) in +936C > T SNP was significantly related to Dukes' stage and tumor location (Table IV); 27.4% of localized tumors showed a T allele whereas 72.6% of these tumors only carried C allele. In 40% of stage C tumors a T allele was detected, and only in 11.9% of stage D tumors this allele was found (p = 0.01). With regard to location, T allele was present in 12.8% of proximal colon tumors, and it was absent in 87.2% of this kind of tumors (p = 0.03).
The median follow-up period was 72 months (6 years); the interquartile range was 27-63 months. In our patient population OS at 6 years was 67%. All survival analyses were adjusted to the median follow-up period. During follow-up, 90 patients died, 70 as a consequence of neoplasia. One patient was lost to follow-up.
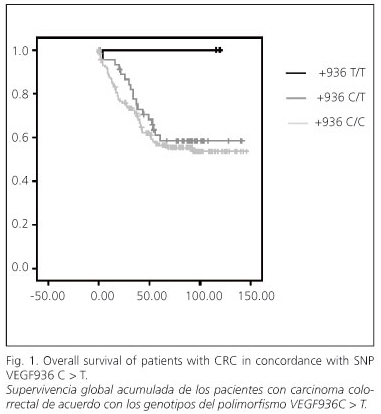
In the univariate analysis no significant differences in OS were found for any of the studied polymorphisms. However, for the +936C > T SNP, patients who showed a T/T genotype had 100% OS after 6 years, patients with a C/T genotype had 59.8% OS, and patients with a C/C genotype had 55.5% OS. This difference was not statistically significant due to the low frequency of the T/T genotype, as only two patients showed it (Fig. 1). The study of OS and other clinicopathological variables is shown in table V. Patients with localized tumors had 83.1% OS against patients with stages C and D, who had 49.3 and 8.0% OS, respectively (p < 0.001).
A stratified analysis of OS was performed using the clinicopathological variables which were statistically significant in the previous analysis. Any of the variables showed significant differences, probably due to the population size with infrequent alleles.
In our population, DFS after 6 years was 70%. During follow-up, recurrence was detected in 36 patients. Recurrence was locoregional in 11 (30.5% patients), and distant recurrence was confirmed in 25 (69.5%) patients.
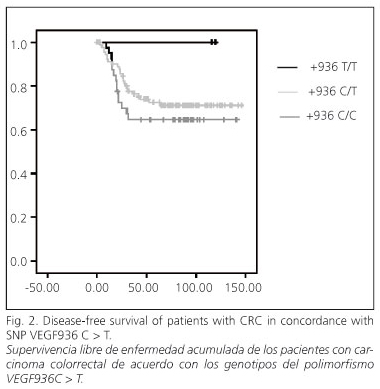
In the univariate analysis no significant differences in SLE were seen according to the three SNPs studied (Fig. 2). The analysis of SLE and the clinicopathological variables is summarized in table V. In women, SLE was 77.9%, and in men it was 63.6%, significantly lower (p = 0.05). According to Dukes' stage SLE was 100% in stages A and B, 77% in stage C, and 48.8% in stage D (p = 0.003).
No significant differences were found in the stratified analysis of SLE with clinicopathological variables.
Discussion
Angiogenesis is a sequence of processes starting with vessel dilatation and pericyte recruitment in pre-existing vessels, followed by endothelial cell proliferation, formation of new vessels, and recruitment of perivascular cells. Therefore, angiogenesis is a key element in tumor development, invasion and metastasis. VEGF is one of the growth factors involved directly in the control of angiogenesis (15,16).
The VEGF gene is located in chromosome 6p12 and is composed of eight exons and seven introns (6). It is a polymorphic gene with more than 15 SNPs described (7,17). Some of these polymorphisms (-2578C > A, -1154G > A, -634G > C and +936C > T) have been related to protein expression (7,10). In this study we have analyzed the -2578C > A, -1154G > A and +936C > T VEGF SNPs and their relation with clinicopathological variables and survival in a population of 177 patients operated on for colorectal carcinoma.
-2578C > A SNP distribution in our population was 36.6% CC genotype, 36.7% CA genotype and 37.7% AA genotype. This distribution is similar to the one found by Lemos et al. (18) for this SNP in a control group of healthy subjects. However, the distribution of the -1154G > A SNP they found in the group of patients shows some differences with our data. The frequency of allele A in our population is higher than in the study of Lemos et al. (22 vs. 7%) (18). The distribution of allele +936C > T SNP in our group of patients was very similar to the frequency observed by other authors in a group of healthy subjects (19,20).
Different studies have demonstrated that VEGF gene expression is influenced by SNPs occurring in this gene. In this manner, -2578C > A and -1154C > G have been related to increased VEGF protein expression (11,12) while the presence of a T allele in the +936 C > T SNP correlated with lower levels of this protein (10). In a meta-analysis published by De Guetz et al., which included 27 studies, they found that VEGF overexpression is significantly correlated with lower OS and increased risk of recurrence (21).
We found a significant relation of T allele in 936C > T SNP and Dukes' stage. This allele exhibits low frequency in stage D and is related to early stages of the disease. In the analysis of OS, patients with the +936T/T genotype showed 100% OS. However, other authors have associated this genotype to advanced stages and distant metastases (22), and the presence of allele T has been related to shorter survival (23,24).
Our findings are based on the analysis of 177 patients operated on for CRC by a single surgeon. The prevalence of genotype +936T/T in our population was 1.1%, therefore it would be necessary to increase our population size to confirm these data. In case we could confirm our findings, the analysis of the +936C > T SNP would be helpful in the follow-up and management of these patients.
References
1. Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular specific growth factors and blood vessel formation. Nature 2000; 407: 242-8. [ Links ]
2. Roy H, Bhardawai S, Yla-Herttuala S. Biology of vascular endothelial growth factors. Febs Lett 2006; 580: 2879-87. [ Links ]
3. Yamamori M, Sakaeda T, Nakamura T, Okamura N, Tamura T, Aoyama N, et al. Association of VEGF genotype with mRNA level in colorectal adenocarcinomas. Biochem Biophys Res Commun 2004; 325: 144-50. [ Links ]
4. Lu H, Shu Z-O, Cui Y, Kataoka N, Wen W, Cai Q, et al. Association of genetic polymorphims in the VEGF gene with breast cancer survival. Cancer Res 2005; 65: 5015-19. [ Links ]
5. Tzanakis N, Gazouli M, Rallis G, Giannopoulos G, Papaconstantinou I, Theodoropoulos G, et al. Vascular endothelial growth factor polymorphisms in gastric cancer development, prognosis and survival. J Surg Oncol 2006; 94: 624-30. [ Links ]
6. Vincenti V, Cassano C, Rocchi M, Persico G. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation 1996; 93: 1493-5. [ Links ]
7. Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: correlation with variation in VEGF protein production. Cytokine 2000; 12: 1232-5. [ Links ]
8. Lin CC, Wu HC, Tsai FJ, Chen HY, Chen WC. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for prostate cancer. Urology 2003; 62: 374-7. [ Links ]
9. Ku KT, Wan L, Peng HC, Tsai MH, Tsai CH, Tasai FJ. Vascular endothelial growth factor gene-460 C/T polymorphism is a biomarker for oral cancer. Oral Oncol 2005; 41: 497-502. [ Links ]
10. Renner W, Kotschan S, Hoffman C, Obermayer-Pietsch B, Pilger E. A common 936 C/T mutation in the gene for vascular endothelial growth factor is associated with vascular endothelial factor plasma levels. J Vasc Res 2000; 37: 443-8. [ Links ]
11. Koukourakis MI, Papazoglou D, Giatromanolaki A, Bougioukas G, Maltezos E, Sivridis E. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer 2004; 46: 293-8. [ Links ]
12. Yang B, Cross DF, Ollerenshaw M, Millward BA, Demaine AG. Polymorphisms of the vascular endothelial growth factor and susceptibility to diabetic microvascular complications in patitients with type 1 diabetes mellitus. J Diabetes Complications 2003; 17: 1-6. [ Links ]
13. Cerdán J. Seguimiento de los pacientes intervenidos de cáncer colorrectal. Rev Cancer 1997; 11: 32-41. [ Links ]
14. Schneider BP, Radovich M, Sledge GW, Robarge JD, Li L, Storniolo AM, et al. Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat 2008; 111(1): 157-63. [ Links ]
15. Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med 2003; 9: 669-76. [ Links ]
16. Flolkman J. Angiogenesis in cancer, vascular, rheumatoid and ther disease. Nat Med 1995; 1: 27-31. [ Links ]
17. Brogan IJ, Khan N, Isaac K, Hutchison JA, Pravica V, Hutchinson IV. Novel polymorphisms in the promoter and 50 UTR regions of the human vascular endothelial growth factor gene. Hum Immunol 1999; 60: 1245-9. [ Links ]
18. Lemos FB, Mol WM. The beneficial effects of recipient derived vascular endothelial growth factor on graft survival alter kidney transplantation. Transplantation 2005; 79(9): 1221-5. [ Links ]
19. Krippl P, Langsenlehner U, Renner W, Yazdani-Biuki B, Wolf G, Wascher TC, et al. A common 936 C/T gene polymorphism of vascular endothelial growth factor is associated with decreased breast cancer risk. Int J Cancer 2003; 106: 468-71. [ Links ]
20. Wu G, Hasenberg T, Magdeburg R, Bönninghoff R, Sturm J, Keese M. Association between EGF, TGF-B1, VEGF gene polymorphism and colorectal cancer. World J Surg 2009; 33: 124-9. [ Links ]
21. Des Guetz G, Uzzan B, Nicolas P, Cucherat M, Morere JF, Benamouzig R, et al. Microvessel density and VEGF expression are prognostic factors in colorrectal cancer. Meta-analysis of the literature. Br J Cancer 2006; 91(12): 1823-32. [ Links ]
22. Chae YS, Kim JG, Sohn SK, Cho YY, Ahn BM, Moon JH, et al. Association of vascular endothelial growth factor gene polymorphisms wit susceptibility and clinicopathologic characteristics of colorectal cancer. J Korean Med Sci 2008; 23: 421-7. [ Links ]
23. Kim JG, Chae YS, Sohn SK, Cho YY, Moon JH, Park JY, et al. Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with colorectal cancer. Clin Cancer Res 2008; 14(1): 62-6. [ Links ]
24. Dassoulas K, Gazouli M, Rizos S, Theodoropoulos G, Christoni Z, Nikiteas N, et al. Common polymorphisms in the vascular endothelial growth factor gene and colorectal cancer development, prognosis and survival. Molecular Carcinogenesis 2009; 48: 563-9. [ Links ]
![]() Correspondence:
Correspondence:
Mª Luisa Maestro de las Casas.
Servicio de Análisis Clínicos.
Hospital Clínico San Carlos.
C/ Martín Lagos, s/n. 28040 Madrid.
e-mail: mmaestro.hcsc@salud.madrid.org
Received: 25-08-09.
Accepted: 13-10-09.











 texto en
texto en