Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
Revista Española de Enfermedades Digestivas
versión impresa ISSN 1130-0108
Rev. esp. enferm. dig. vol.108 no.9 Madrid sep. 2016
https://dx.doi.org/10.17235/reed.2016.4261/2016
DOI: 10.17235/reed.2016.4261/2016
ORIGINAL PAPERS
Gastric microbiota and carcinogenesis: the role of non-Helicobacter pylori bacteria - A systematic review
Emanuel Dias-Jácome1, Diogo Libânio2, Marta Borges-Canha3, Ana Galaghar4 and Pedro Pimentel-Nunes1,2,3
1CINTESIS - Center for Health Technology and Services Research. Faculty of Medicine. University of Porto. Porto, Portugal.
2Gastroenterology Department. Oncology Portuguese Institute of Porto. Porto, Portugal.
3Department of Physiology and Cardiothoracic Surgery. Cardiovascular Research & Development Unit. Faculty of Medicine. University of Porto. Porto, Portugal.
4Pathology Department. Oncology Portuguese Institute of Porto. Porto, Portugal.
ABSTRACT
Background and aim: Helicobacter pylori is the strongest risk factor for gastric cancer. However, recent advances in DNA sequencing technology have revealed a complex microbial community in the stomach that could also contribute to the development of gastric cancer. The aim of this study was to present recent scientific evidence regarding the role of non-Helicobacter pylori bacteria in gastric carcinogenesis.
Methods: A systematic review of original articles published in PubMed in the last ten years related to gastric microbiota and gastric cancer in humans was performed.
Results: Thirteen original articles were included. The constitution of gastric microbiota appears to be significantly affected by gastric cancer and premalignant lesions. In fact, differences in gastric microbiota have been documented, depending on Helicobacter pylori status and gastric conditions, such as non-atrophic gastritis, intestinal metaplasia and cancer. Gastric carcinogenesis can be associated with an increase in many bacteria (such as Lactobacillus coleohominis, Klebsiella pneumoniae or Acinetobacter baumannii) as well as decrease in others (such as Porphyromonas spp, Neisseria spp, Prevotella pallens or Streptococcus sinensis). However, there is no conclusive data that confirms if these changes in microbiota are a cause or consequence of the process of carcinogenesis.
Conclusions: Even though there is limited evidence in humans, microbiota differences between normal individuals, pre-malignant lesions and gastric cancer could suggest a progressive shift in the constitution of gastric microbiota in carcinogenesis, possibly resulting from a complex cross-talk between gastric microbiota and Helicobacter pylori. However, further studies are needed to elucidate the specific role (if any) of different microorganisms.
Key words: Stomach. Microbiota. Bacteria. Carcinogenesis. Helicobacter pylori.
Introduction
Human beings are colonized by a myriad of microorganisms that interact with each other and with the host in a symbiotic manner, constituting an integrated and functional ecosystem (microbiota) that can influence health as well as disease (1,2). In particular, gastrointestinal microbiota can modulate cancer risk within the gastrointestinal tract (3). Most research today has focused on colon microbiota and its relationship with colorectal cancer (4) and has rarely examined gastric microbiota. Historically, the stomach was considered as a sterile organ due to acid production.
The discovery of Helicobacter pylori (H. pylori) demolished this belief and, for many years, this bacterium was thought to be the only bacteria able to survive in the stomach. However, recent evidence suggests that other bacteria besides H. pylori flourish despite the hostile environment, and gastric microbiota is, actually, more diverse and complex than once thought. Its composition can be shaped by the host physiology as well as by external factors such as diet, H. pylori infection, enteral feeding, proton pump inhibitors, antibiotics and disease (5).
Correa's model of gastric carcinogenesis postulated that chronic H. pylori infection of the gastric mucosa progresses through the stages of chronic active gastritis, atrophic gastritis, intestinal metaplasia, dysplasia and, ultimately, gastric cancer (6). Today, H. pylori is clearly identified as the strongest risk factor for gastric cancer (7,8), the fourth most common malignancy and the second leading cause of cancer-related mortality worldwide (9,10).
However, only 1-2% of infected people develop gastric cancer (11), and its pathogenic mechanisms are unclear. It is possible that gastric microbiota constitutes the link between H. pylori and gastric carcinogenesis, when other bacteria colonize the stomach under conditions of decreased acidity (such as chronic atrophic gastritis), creating reactive oxygen and nitrogen species and modulating inflammatory responses (12,13). Therefore, it is important to unravel the significance of specific microbiota to determine which ones are causative of gastric cancer and which are present merely as a consequence in order to develop novel prevention and treatment strategies.
This systematic review aims to provide an update of recent findings related to the role of gastric microbiota in gastric carcinogenesis, focusing on microorganisms besides H. pylori to understand how the constitution of microbiota can influence cancer risk and how it is affected by H. pylori status, possibly explaining its pathogenesis.
Methods
A comprehensive search was performed using PubMed and the following query (date of last search 5th August 2015): ("microbiome" OR "microbiota" OR "microflora" OR "bacterial flora" OR "bacterial community") AND ("gastric" OR "stomach" OR "upper digestive tract" OR "upper gastrointestinal tract") AND ("cancer" OR "neoplasia" OR "malignancy" OR "tumor" OR "carcinoma" OR "adenocarcinoma" OR "lymphoma" OR "premalignancy" OR "premalignant" OR "tumorigenesis" OR "carcinogenesis" OR "intestinal metaplasia" OR "atrophic gastritis" OR "Helicobacter pylori" OR "H. pylori").
In the first phase, titles and abstracts were screened by two reviewers, according to specific criteria defined in order to guide this systematic review. Any disagreement was resolved by a third independent reviewer. We included studies presenting information about the role of gastric microbiota beyond H. pylori in gastric carcinogenesis or presenting information about the influence of H. pylori in the constitution of gastric microbiota. The exclusion criteria were: articles published before 2005 (because the constitution of gastric microbiota has been defined more accurately approximately since this year, as previous studies were based on Methods with limitations in sensitivity and coverage when compared with more recent approaches); articles not written in English, Portuguese or Spanish; studies performed in animals; studies performed in cell culture models; non-original studies (reviews and Case Reports).
In the next phase, full-texts of the selected References were assessed and References of included studies were checked to identify additional articles.
Data about bacterial species studied and Results obtained were extracted from each article, mentioning Methods and main limitations when relevant. The information was then summarised and organized in the present systematic review.
Results
Characteristics of included studies
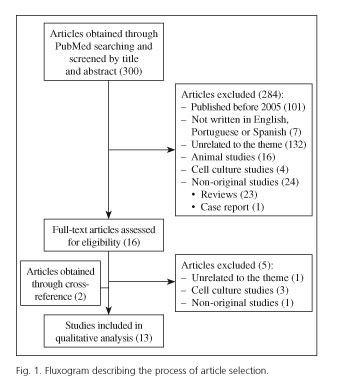
A total of 300 articles were obtained from the search of the database. After screening by title and abstract, 16 studies were selected for full-text reading, after which, 5 studies were further excluded and 2 studies included by cross-reference. Thirteen articles were finally included (Fig. 1). Their general characteristics are summarized in table I.
Influence of normal gastric microbiota by H. pylori
Characterization of gastric microbiota in subjects with and without H. pylori could provide new insight into the mechanisms behind a hypothetical cross-talk between this bacterium and the remaining bacterial community and its possible link with gastric carcinogenesis. However, the effect of H. pylori on the microbiota of the human stomach has been examined in very few studies.
H. pylori and gastric microbiota in adults
In 2006, Bik et al. characterized bacterial diversity within the human gastric mucosa using a small subunit 16S rDNA clone library approach. This group analyzed 1,833 sequences generated by broad-range bacterial PCR from gastric biopsy samples of 23 individuals. A diverse gastric bacterial community of 128 phylotypes was dominated by 5 major phyla: Proteobacteria (952 clones, 32 phylotypes), Firmicutes (464 clones, 36 phylotypes), Bacteroidetes (193 clones, 35 phylotypes), Actinobacteria (164 clones, 12 phylotypes), and Fusobacteria (56 clones, 10 phylotypes). H. pylori was the only member of the genus Helicobacter found; the sequences were found in 19 of the 23 samples analyzed and constituted 42% of all sequences analyzed (777 clones), making this the most abundant genus followed by Streptococcus (299 clones) and Prevotella (139 clones). Interestingly, its presence did not affect significantly the composition of gastric microbiota, although H. pylori-positive subjects demonstrated a relative lack of non-Proteobacteria phylotypes, especially Bacteroidetes (p > 0.05) (14).
A similar constitution of gastric microbiota was reported by Li et al., who analyzed gastric microbiota of 10 subjects without H. pylori infection by cloning and sequencing 16S rRNA. A total of 1,223 non-H.pylori sequences were obtained and a diverse bacterial community of 133 phylotypes was dominated by the same 5 phyla: the most abundant was Firmicutes (383 clones), closely followed by Proteobacteria (345 clones), while Bacteroidetes, Actinobacteria and Fusobacteria were less abundant. The most common genera were: Streptococcus (254 clones), Prevotella (243 clones), Neisseria (175 clones), Haemophilus (122 clones) and Porphyromonas (68 clones). All of them constituted 70.5% of all microbial clones (15). However, when only H. pylori-negative subjects were included, the impact of H. pylori in the constitution of microbiota was not evaluated.
Although these two studies analyzed two geographically (Hong Kong vs. California) and ethnically (Chinese vs. Caucasian, Hispanic and African American) distinct populations, the overall microbiota constitution was surprisingly similar. Both identified approximately 130 phylotypes from the same dominant phyla (Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria and Fusobacteria) and the two most abundant genera (Streptococcus and Prevotella) were also identical (14,15).
However, contradicting findings have been reported and other studies suggest that H. pylori can actually change the composition of gastric microbiota (16,17).
In 2008, Andersson et al. analyzed gastric biopsy samples from six Swedish healthy individuals (three H. pylori-infected and three uninfected) by tagged 454-pyrosequencing, and found a greater gastric diversity in H. pylori negative patients (262 phylotypes) than in H. pylori positive patients (33 phylotypes) (16).
In 2011, Maldonado-Contreras et al. characterized the structure of the human gastric bacterial community in relation to H. pylori status in 12 adult patients (10 Amerindians and 2 non-Amerindians) using a high-density 16S rRNA gene microarray. A total of 44 bacterial phyla were detected in an uneven ecosystem, with a strong dominance of only four phyla: Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes (in descending order). Positive H. pylori status was significantly associated with increased relative abundance of non-Helicobacter bacteria from the Proteobacteria, Spirochetes and Acidobacteria, and with decreased abundance of Actinobacteria, Bacteroidetes and Firmicutes. In fact, about 28% of the total variance in the gastric microbiota of the 12 subjects was explained by H. pylori status, although a larger fraction (44%) was related to the ethnicity of the subjects, stressing the need for conducting larger studies of different human populations (17).
More recently, other studies based on bacterial cultures analyzed by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS) allow high accuracy and rapid analysis of a large number of samples (18,19).
Hu et al. adopted this approach to analyze 103 gastric biopsy specimens from H. pylori infected patients presenting with dyspeptic symptoms (caused by gastritis, gastric ulcer, duodenal ulcer, reflux esophagitis and non-ulcer dyspepsia). A total of 201 non-H. pylori bacterial isolates were cultured from samples from 67 (65%) patients and all the isolates were identified using MALDI-TOF MS. Overall, 18 non-H. pylori bacterial genera (43 species) were isolated, mostly gram-positive bacteria (73.8%). The dominant species were: Neisseria flavescens (13.7%), Streptococcus salivarius (9.5%), Rothia mucilaginosa (8.9%) and Streptococcus pneumoniae (6.6%). This study demonstrates a high prevalence of the non-H. pylori bacteria concurrent with H. pylori infection, indicating that upper gastrointestinal disorders may enable non-H. pylori bacteria to survive and colonize the stomach (18). However, this approach excludes non-cultivable bacteria and does not include H. pylori-uninfected subjects, so it does not evaluate its impact in the constitution of gastric microbiota.
In an attempt to overcome this limitation, Khosravi et al. adopted a similar approach to compare culturable gastric microbiota in H. pylori positive and negative gastric disease patients in a large sample of 215 Malaysian patients (131 H. pylori-positive and 84 H. pylori-negative). Notably, H. pylori colonization did not significantly affect the constitution of the human gastric microbiota (19), consistent with the Results reported by Bik et al. (14). Similarly, Maldonado-Contreras et al. did not report a variation in the absolute number of non-H. pylori bacteria present but found that H. pylori resulted in differences in the relative abundance of several phyla (17).
Predominant phyla were Firmicutes (270 colonies from 208 positive biopsies) and Proteobacteria (220 colonies from 160 biopsies) while Actinobacteria were rare (53 colonies from 43 biopsies). At the genus level, Streptococcus were predominant (121 positive biopsies) followed by Neisseria (44 positive biopsies), Klebsiella (42 positive biopsies) and Lactobacillus (42 positive biopsies) (19). These Results are consistent with previous studies reporting that Proteobacteria, Firmicutes, Actinobacteria and Bacteroidetes are the predominant phyla in human gastric microbiota (14-17), overcoming the important limitation of the small number of human samples (6-23 individuals) in these studies. Nevertheless, the approach adopted by Khosravi et al. still excludes non-cultivable bacteria.
Eun et al. also analyzed the gastric microbiota profile according to H. pylori status using high-throughput 16S rRNA sequencing, in biopsy samples collected from 31 patients (11 with noncardia gastric cancer, 10 with intestinal metaplasia, and 10 with chronic gastritis) during endoscopy. The relative abundance of the families Bradyhizobiaceae, Caulobacteraceae, Lactobacillaceae, and Burkholderiaceae was significantly higher in H. pylori-negative patients, whereas the relative abundance of the Helicobacteriaceae family was significantly higher in H. pylori-positive patients (20).
H. pylori and gastric microbiota in children
Information about gastric microbiota in children is even more limited; to date, only Kato et al. addressed this question by comparing gastric microbiota in 10 children and 10 adults presenting with dyspeptic symptoms in relation to H. pylori status (5 H. pylori-infected and 5 uninfected in each group) using both culture and RT-PCR techniques. An overall colonization rate of non-Helicobacter bacteria in gastric mucosa was higher in adults (100%) than in children (10%) (p < 0.001). Also, there was no significant difference between adults with H. pylori infection and those without (p = 0.056). This demonstrates that, regardless of H. pylori status, bacteria reside on the gastric mucosa of adults, although such microorganisms rarely colonize the stomach of children (21).
The role of microbiota in the development of gastric cancer
The very few studies that compare the constitution of gastric microbiota between gastric cancer patients and controls, in order to establish whether there are significant differences that could support a possible causal role for these bacteria, are analyzed in this section.
Global gastric microbiota diversity and gastric cancer
Avilés-Jiménez et al. analyzed the constitution of gastric microbiota in five patients, each with non-atrophic gastritis, intestinal metaplasia and gastric cancer, and observed low gastric microbiota diversity in all three disease groups: two phyla, Firmicutes and Proteobacteria, represented almost 70% of microbial community while 9 families represented on average 55.6% of each sample's operational taxonomic units (OTU), with Lachnospiraceae and Streptococacceae representing over 20% of the microbiota. In addition, although significant differences in bacterial diversity were noted between non-atrophic gastritis and gastric cancer (p = 0.004) but not between intestinal metaplasia and any other group, a decreasing trend from non-atrophic gastritis to intestinal metaplasia to gastric cancer was observed (22).
A cross-sectional study involving 720 Chinese healthy volunteers performed by Yu et al. supports the idea that decreased gastric microbial diversity could favor the development of gastric cancer. This study found a linear relationship between microbial richness and pepsinogen I/pepsinogen II ratio. Therefore, lower gastric microbial richness is associated with a lower pepsinogen I/pepsinogen II ratio, which is a marker for chronic atrophic gastritis, a cancer-predisposing condition in the stomach (23).
However, Eun et al. compared gastric microbiota between patients with gastric cancer, intestinal metaplasia and chronic gastritis and reported that the evenness and diversity of gastric microbiota was significantly increased in gastric cancer (20).
Specific changes in microbiota possibly related to gastric carcinogenesis
Dicksved et al. contributed the first DNA-based description of a complex gastric bacterial community in individuals with gastric cancer with potential as a maintainer or bystander in the progression of neoplasia. This group compared the gastric microbiota of 10 patients with gastric cancer with that of 5 dyspeptic controls with normal mucosa using terminal restriction fragment length polymorphism (T-RFLP) in combination with 16S rRNA gene cloning and sequencing. Sequencing of 140 clones revealed 102 phylotypes, with representatives from five bacterial phyla: Firmicutes (60%), Bacteroidetes (11%), Actinobacteria (7%), Proteobacteria (6%) and Fusobacteria (3%). The abundance of H. pylori was very low and gastric cancer microbiota was instead dominated by different species of the genera Streptococcus, Lactobacillus, Veillonella and Prevotella. Remarkably, the constitution of gastric microbiota was not significantly different between gastric cancer patients and controls (p = 0.12) (24).
In contrast to the aforementioned Results, Eun et al. report significant differences in the composition and diversity of gastric microbiota among patients with gastric cancer, intestinal metaplasia and chronic gastritis, especially in the Helicobacter-dominant group. The gastric cancer group showed a significant decrease in the Epsilonproteobacteria class and Helicobacteriaceae family, while the Bacilli class and Streptococacceae family increased significantly (20).
In addition, significant differences in the gastric microbiota profile between non-atrophic gastritis, intestinal metaplasia and intestinal type gastric cancer were recently reported by Avilés-Jiménez et al. Further analyses revealed 27 operational taxonomic units (OTU) with a difference in abundance significant across groups. From the 12 OTU with the most significant p-values (as selected by the authors), 10 displayed a clear decreasing trend from non-atrophic gastritis to intestinal metaplasia to gastric cancer (2 TM7, 2 Porphyromonas spp, 1 Neisseria spp, 1 Haemophilus spp, 1 Bulleidia spp, Prevotella pallens, Streptococcus sinensis and Bergeriella denitrificans), while 2 revealed an increasing trend (Lactobacillus coleohominis and Lachnospiraceae) (22).
The approach taken by Khosravi et al. also aimed to provide an insight into the contribution of disease to the constitution of the gastric microbiota. The gastric microbiota was compared between patients with different gastric diseases, including non-ulcer dyspepsia (87.6%), peptic ulcer disease (10.2%) and gastric cancer (3.7%). Interestingly, the prevalence of Klebsiella pneumoniae and Acinetobacter baumannii in gastric cancer patients was significantly higher than that in the non-ulcer dyspepsia and peptic ulcer disease groups (50% vs 18.9% and 9.10%; 25% vs. 3.2% and 4.5%, respectively). Nevertheless, these Results should be considered with caution because of the low number of gastric cancer patients compared with the two other groups (19).
The presence of Bifidobacteriaceae in the hypochlorhydria stomach of 13 patients with atrophic gastritis and 10 patients receiving omeprazole was recently studied by Matarelli et al. The presence of Bifidobacteriaceae correlated significantly only in the omeprazole patients. However, bacteria belonging to the Actinomycetales order were also present and correlated significantly with both conditions (p < 0.01) (25). It is surprising that previous studies on gastric cancer and chronic atrophic gastritis failed to detect these bacteria (20,22,24). The significance of this atypical microbiota colonization remains to be studied.
The presence of Helicobacter DNA was investigated by Han et al. using a Helicobacter species-specific 16S rRNA PCR amplification and pyrosequencing analysis in 51 resected gastric adenocarcinomas. H. pylori was present in 47 (92.2%) of 51 samples. In the four of H. pylori-negative cases, two cases of Helicobacter cinaedi were detected, as well as one case of Helicobacter mustelae and one case of Campylobacter hyointestinalis. However, their role in the pathogenesis of gastric adenocarcinoma remains unclear (26).
Taken together, these Results demonstrate that there are significant differences in the constitution of gastric microbiota between the sequential steps of the cascade of carcinogenesis, such as non-atrophic gastritis, intestinal metaplasia and gastric cancer. These findings suggest that there could be a progressive shift in the constitution of the gastric microbiota during the progression of pre-malignant lesions to gastric cancer and that gastric carcinogenesis could be associated with an increase in many bacteria, as well as a decrease in others, as summarized in table II.
It is interesting to note that, globally, there is a decrease in bacteria belonging to the phyla Proteobacteria (Neisseria spp [22], Haemophilus spp [22], Bergeriella denitrificans [22], Epsilonproteobacteria [20] and Helicobacteriaceae [20]) and Bacteroidetes (Porphyromonas spp [22] and Prevotella pallens [22]), whereas bacteria belonging to the phyla Firmicutes are either increased (Streptococacceae [20], Lachnospiraceae [22] and Lactobacillus coleohominis [22]) or decreased (Streptococcus sinensis [22]). Klebsiella pneumoniae and Acinetobacter baumannii are two notable exceptions, as they belong to the phylum Proteobacteria, although they tend to increase with gastric carcinogenesis (19). It is intriguing that one study found that Streptococcus sinensis was associated with a decreasing trend from premalignant lesions to gastric cancer (22), whereas another study reported that its family, Streptococacceae, was increased in gastric cancer compared with intestinal metaplasia and chronic gastritis (20).
Bacteria belonging to the other two dominant phyla, Actinobacteria and Fusobacteria, appear to play a less significant role in the process of carcinogenesis, as only one study reported a significant increase in bacteria belonging to the phyla Actinobacteria (Actinomycetales [25]) in atrophic gastritis, whereas there are no studies reporting significant variations involving Fusobacteria.
Discussion
The present systematic review shows that, even though there is little evidence in humans, there could be a progressive shift in the constitution of gastric microbiota as well as the progression from premalignant lesions to gastric cancer, and, possibly, these microorganisms play an important (as yet undetermined) role in gastric carcinogenesis, beyond the well-known influence of H. pylori.
Studies regarding the role of H. pylori in the constitution of gastric microbiota have provided conflicting Results, which could be related to the Methods used. In fact, some of the included studies did not find significant differences using cloning (14) and culture-based approaches (19,21), whereas other studies report significant changes in gastric microbiota according to H. pylori status based on DNA microarrays (17) or 454-pyrosequencing (16), which allow the study of a wider range of bacteria, thus providing a more complete view of the picture. Animal studies have also yielded contradicting findings, as H. pylori infection altered significantly the constitution of gastric microbiota in a BALB/c mice model (27) and in Mongolian gerbils (28), but it does not significantly alter the murine gastric microbiota (29).
Dicksved et al. contributed the first DNA-based description of a complex gastric microbiota composition in individuals with gastric cancer although it did not differ significantly between gastric cancer patients and controls. However, this study has considerable limitations, including the small sample size and lack of the most current high-throughput sequencing technology (24). In fact, more recent studies reported significant differences in the constitution of gastric microbiota between patients with gastric cancer and premalignant lesions (20,22), which could be related to the method used: the study conducted by Dicksved et al. was based on cloning, sequencing and T-RFLP of 16S rRNA (24), which has important limitations in sensitivity and coverage compared with more recently developed 16S rRNA sequencing techniques, including G3 PhyloChip (22) or 454 GS FLX Titanium (20).
It is also interesting that some studies suggest that gastric carcinogenesis is related to low gastric microbial diversity (22,23), which contrasts with the increased diversity in gastric cancer patients reported by Eun et al. (20). This could be partially explained by the fact that Yu et al. used an array primarily designed to assess oral microbiota (HOMIM), possibly excluding some bacteria from the stomach, and the lower microbial richness could actually reflect their increased abundance (23). Nevertheless, this is not the case of the Results reported by Avilés-Jiménez et al., who used a G3 PhyloChip array (22), while Eun et al. used a 454 high-throughput sequencer (20). Both approaches are reliable to characterize gastric microbiota and this discrepancy is intriguing.
Other reviews (30-34) and recent studies in animal models (35-37) strongly support an important role for gastric microbiota in gastric carcinogenesis. In fact, the colonization of insulin-gastrin (INS-GAS) mice, a mouse model of gastric cancer, by a complex gastric microbiota not only increased the incidence of gastric intraepithelial neoplasia (36), but also accelerated its development when compared with those infected by H. pylori alone (35). In addition, antibiotic treatments had the opposite effect, delaying significantly the development of gastric intraepithelial neoplasia (37).
Taking all these Results together, one hypothesis is that gastric carcinogenesis could result from the interaction between H. pylori and gastric microbiota. H. pylori may be more important in the early stages of gastric carcinogenesis, as a trigger for the development of chronic atrophic gastritis, rather than as a direct inducer of gastric cancer. Instead, the decreased acidity secondary to mucosal atrophy would lead to changes in the constitution of gastric microbiota that could play important roles in the later stages of gastric carcinogenesis, inducing progression from chronic atrophic gastritis to intestinal metaplasia and, finally, to gastric cancer (Fig. 2).
The link between H. pylori and gastric microbiota in the pathogenesis of gastric cancer can be related to Toll-like receptors. In fact, there is evidence that increasing activation of these receptors, initially by H. pylori and later potentially by other microorganisms, may play an important role in gastric carcinogenesis (38).
Although our knowledge about the role of gastric microbiota in gastric carcinogenesis is only in the beginning, it looks clear that this is an area of intensive research and great future potential. In fact, since our timeline (August 2015), two more original studies have been published. Jo et al. analyzed gastric microbiota by 454-pyrosequencing of the 16S rRNA gene and reported that the number of nitrate-reducing bacteria other than H. pylori was twofold higher in the cancer group compared with the control group, but it did not reach statistical significance (39). Another recent study, performed by Yang et al., compared gastric microbiota between two Colombian populations with very different incidences of gastric cancer (Tuquerres have a 25-fold higher risk than the Tumaco) in spite of a similar H. pylori prevalence. Interestingly, these authors found that the gastric microbiota had a significant correlation with the town of origin, with Leptotrichia wadei and Veilonella sp being significantly more common in Tuquerres and Staphylococcus sp significantly more abundant in Tumaco (40). These Results demonstrate that this is an area of intensive research and our knowledge about the role of gastric microbiota in gastric carcinogenesis will certainly progress in the near future. We believe that this systematic review provides an important starting point for future research in this area.
Perhaps further knowledge about the role of specific microorganisms in the progression of gastric premalignant lesions would allow the development of specific microbes and microbiota-based biomarkers for diagnostic and therapeutic purposes. Certain microorganisms with anti-H. pylori activity, like Lactobacillus (41,42) or Saccharomyces boulardii (43), could be used as a novel probiotic approach for H. pylori eradication, overcoming antibiotic-associated problems like adverse effects or resistance. Also, if a gradual shift in the gastric microbiota profile from non-atrophic gastritis to intestinal metaplasia, and then to intestinal type gastric cancer, is confirmed, perhaps a novel screening method based on searching for specific gastric microbiota profiles could be developed, allowing us to detect premalignant lesions.
This systematic review faces some limitations. There are few studies about the role of gastric microbiota in human patients and most involve a limited number of subjects and follow a cross-sectional design. Also, most studies included do not make a clear distinction between diffuse and intestinal types of gastric cancer. More studies in large prospective cohorts are needed to confirm how the human gastric microbiota may affect the development of gastric cancer.
Nevertheless, this study, to our knowledge, is the first systematic review summarizing the evidence regarding the influence of non-Helicobacter bacteria in the process of gastric carcinogenesis. However, current evidence does not allow us to make specific Conclusions about the specific role of other bacteria in the development of gastric cancer, with many potentially being only innocent bystanders.
In conclusion, the interaction between H. pylori and gastric microbiota may be a crucial step in gastric carcinogenesis. Microbiota differences between healthy individuals, pre-malignant lesions and gastric cancer have been described, and it could suggest a progressive shift in the constitution of gastric microbiota in carcinogenesis. However, the identities of specific microorganisms involved in these processes remain largely unknown and further studies clarifying their carcinogenic mechanisms are needed.
References
1. Cho I, Blaser MJ. The human microbiome: At the interface of health and disease. Nat Rev Genet 2012;13(4):260-70. DOI: 10.1038/nrg3182. [ Links ]
2. Plottel CS, Blaser MJ. Microbiome and malignancy. Cell Host Microbe 2011;10(4):324-35. DOI: 10.1016/j.chom.2011.10.003. [ Links ]
3. Abreu MT, Peek RM Jr. Gastrointestinal malignancy and the microbiome. Gastroenterology 2014;146(6):1534-46.e3. DOI: 10.1053/j.gastro.2014.01.001. [ Links ]
4. Borges-Canha M, Portela-Cidade JP, Dinis-Ribeiro M, et al. Role of colonic microbiota in colorectal carcinogenesis: A systematic review. Rev Esp Enferm Dig 2015;107(11):659-71. DOI: 10.17235/reed.2015.3830/2015. [ Links ]
5. Wu WM, Yang YS, Peng LH. Microbiota in the stomach: New insights. J Dig Dis 2014;15(2):54-61. DOI: 10.1111/1751-2980.12116. [ Links ]
6. Correa P. Human gastric carcinogenesis: A multistep and multifactorial process-First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res 1992;52(24):6735-40. [ Links ]
7. Polk DB, Peek RM Jr. Helicobacter pylori: Gastric cancer and beyond. Nat Rev Cancer 2010;10(6):403-14. DOI: 10.1038/nrc2857. [ Links ]
8. Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 2001;345(11):784-9. DOI: 10.1056/NEJMoa001999. [ Links ]
9. Hu Y, Fang JY, Xiao SD. Can the incidence of gastric cancer be reduced in the new century? J Dig Dis 2013;14(1):11-5. DOI: 10.1111/j.1751-2980.2012.00647.x. [ Links ]
10. Thiel A, Ristimaki A. Gastric cancer: Basic aspects. Helicobacter 2012;17(Suppl 1):26-9. DOI: 10.1111/j.1523-5378.2012.00979.x. [ Links ]
11. Herrera V, Parsonnet J. Helicobacter pylori and gastric adenocarcinoma. Clin Microbiol Infect 2009;15(11):971-6. DOI: 10.1111/j.1469-0691.2009.03031.x. [ Links ]
12. Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut microbes 2013;4(6):505-31. DOI: 10.4161/gmic.26205. [ Links ]
13. Engstrand L, Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol 2013;27(1):39-45. DOI: 10.1016/j.bpg.2013.03.016. [ Links ]
14. Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 2006;103(3):732-7. DOI: 10.1073/pnas.0506655103. [ Links ]
15. Li XX, Wong GL, To KF, et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PloS One 2009;4(11):e7985. DOI: 10.1371/journal.pone.0007985. [ Links ]
16. Andersson AF, Lindberg M, Jakobsson H, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PloS One 2008;3(7):e2836. DOI: 10.1371/journal.pone.0002836. [ Links ]
17. Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J 2011;5(4):574-9. DOI: 10.1038/ismej.2010.149. [ Links ]
18. Hu Y, He LH, Xiao D, et al. Bacterial flora concurrent with Helicobacter pylori in the stomach of patients with upper gastrointestinal diseases. World J Gastroenterol 2012;18(11):1257-61. DOI: 10.3748/wjg.v18.i11.57. [ Links ]
19. Khosravi Y, Dieye Y, Poh BH, et al. Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. ScientificWorldJournal 2014;2014:610421. DOI: 10.1155/2014/610421. [ Links ]
20. Eun CS, Kim BK, Han DS, et al. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing Methods. Helicobacter 2014;19(6):407-16. DOI: 10.1111/hel.12145. [ Links ]
21. Kato S, Fujimura S, Kimura K, et al. Non-Helicobacter bacterial flora rarely develops in the gastric mucosal layer of children. Dig Dis Sci 2006;51(4):641-6. DOI: 10.1007/s10620-006-3185-0. [ Links ]
22. Avilés-Jiménez F, Vázquez-Jiménez F, Medrano-Guzmán R, et al. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep 2014;4:4202. DOI: 10.1038/srep04202. [ Links ]
23. Yu G, Gail MH, Shi J, et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev 2014;23(5):735-41. DOI: 10.1158/1055-9965.EPI-13-0855. [ Links ]
24. Dicksved J, Lindberg M, Rosenquist M, et al. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol 2009;58(Pt 4):509-16. DOI: 10.1099/jmm.0.007302-0. [ Links ]
25. Mattarelli P, Brandi G, Calabrese C, et al. Occurrence of Bifidobacteriaceae in human hypochlorhydria stomach. Microb Ecol Health Dis 2014;25. DOI: 10.3402/mehd.v25.21379. [ Links ]
26. Han HS, Lee KY, Lim SD, et al. Molecular identification of Helicobacter DNA in human gastric adenocarcinoma tissues using Helicobacter species-specific 16S rRNA PCR amplification and pyrosequencing analysis. Oncol Lett 2010;1(3):555-8. [ Links ]
27. Aebischer T, Fischer A, Walduck A, et al. Vaccination prevents Helicobacter pylori-induced alterations of the gastric flora in mice. FEMS Immunol Med Microbiol 2006;46(2):221-9. DOI: 10.1111/rp10.1016-j.femsim.2004.05.008. [ Links ]
28. Osaki T, Matsuki T, Asahara T, et al. Comparative analysis of gastric bacterial microbiota in Mongolian gerbils after long-term infection with Helicobacter pylori. Microb Pathog 2012;53(1):12-8. DOI: 10.1016/j.micpath.2012.03.008. [ Links ]
29. Tan MP, Kaparakis M, Galic M, et al. Chronic Helicobacter pylori infection does not significantly alter the microbiota of the murine stomach. Appl Environ Microbiol 2007;73(3):1010-3. DOI: 10.1128/AEM.01675-06. [ Links ]
30. Brawner KM, Morrow CD, Smith PD. Gastric microbiome and gastric cancer. Cancer J 2014;20(3):211-6. DOI: 10.1097/PPO.0000000000000043. [ Links ]
31. Wang LL, Yu XJ, Zhan SH, et al. Participation of microbiota in the development of gastric cancer. World J Gastroenterol 2014;20(17): 4948-52. DOI: 10.3748/wjg.v20.i17.4948. [ Links ]
32. Wang ZK, Yang YS. Upper gastrointestinal microbiota and digestive diseases. World J Gastroenterol 2013;19(10):1541-50. DOI: 10.3748/wjg.v19.i10.1541. [ Links ]
33. Nardone G, Compare D. The human gastric microbiota: Is it time to rethink the pathogenesis of stomach diseases? United European Gastroenterol J 2015;3(3):255-60. DOI: 10.1177/2050640614566846. [ Links ]
34. Walker MM, Talley NJ. Review article: Bacteria and pathogenesis of disease in the upper gastrointestinal tract-Beyond the era of Helicobacter pylori. Aliment Pharmacol Ther 2014;39(8):767-79. DOI: 10.1111/apt.12666. [ Links ]
35. Lofgren JL, Whary MT, Ge Z, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology 2011;140(1):210-20. DOI: 10.1053/j.gastro.2010.09.048. [ Links ]
36. Lertpiriyapong K, Whary MT, Muthupalani S, et al. Gastric colonization with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut 2014;63(1):54-63. DOI: 10.1136/gutjnl-2013-305178. [ Links ]
37. Lee CW, Rickman B, Rogers AB, et al. Helicobacter pylori eradication prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res 2008;68(9):3540-8. DOI: 10.1158/0008-5472.CAN-07-6786. [ Links ]
38. Pimentel-Nunes P, Goncalves N, Boal-Carvalho I, et al. Helicobacter pylori induces increased expression of Toll-like receptors and decreased Toll-interacting protein in gastric mucosa that persists throughout gastric carcinogenesis. Helicobacter 2013;18(1):22-32. DOI: 10.1111/hel.12008. [ Links ]
39. Jo HJ, Kim J, Kim N, et al. Analysis of gastric microbiota by pyrosequencing: Minor role of bacteria other than Helicobacter pylori in the gastric carcinogenesis. Helicobacter 2016;24. DOI: 10.1111/hel.12293. [ Links ]
40. Yang I, Woltemate S, Piazuelo MB, et al. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep 2016;6:18594. DOI: 10.1038/srep18594. [ Links ]
41. García CA, Henríquez AP, Retamal RC, et al. Probiotic properties of Lactobacillus spp isolated from gastric biopsies of Helicobacter pylori infected and non-infected individuals. Rev Med Chil 2009;137(3):369-76. [ Links ]
42. Chen X, Liu XM, Tian F, et al. Antagonistic activities of lactobacilli against Helicobacter pylori growth and infection in human gastric epithelial cells. J Food Sci 2012;77(1):M9-14. DOI: 10.1111/j.1750-3841.2011.02498.x. [ Links ]
43. Sakarya S, Gunay N. Saccharomyces boulardii expresses neuraminidase activity selective for alpha2,3-linked sialic acid that decreases Helicobacter pylori adhesion to host cells. APMIS 2014;122(10):941-50. DOI: 10.1111/apm.12237. [ Links ]
![]() Correspondence:
Correspondence:
Emanuel Dias-Jácome.
CINTESIS - Center for Health Technology and Services Research.
Faculty of Medicine.
Rua Dr. Plácido da Costa.
4200-450 Porto, Portugal
e-mail: diasj0310@gmail.com
Received: 15-02-2015
Accepted: 13-06-2016