My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista Española de Enfermedades Digestivas
Print version ISSN 1130-0108
Rev. esp. enferm. dig. vol.110 n.7 Madrid Jul. 2018
https://dx.doi.org/10.17235/reed.2018.5269/2017
SPECIAL ARTICLE
Recommendations to report and interpret HLA genetic findings in coeliac disease
1Laboratorio de Investigación en Genética de Enfermedades Complejas. Instituto de Investigación Sanitaria del Hospital Clínico San Carlos (IdISSC). Hospital Clínico San Carlos. Madrid, Spain
2Laboratorio de Genética. Servicio de Análisis Clínicos. Hospital Universitario Río Hortega. Valladolid, Spain
3Grupo de Inmunología de las Mucosas. IBGM. Universidad de Valladolid-CSIC. Valladolid, Spain
INTRODUCTION
Coeliac disease (CD) is a chronic autoimmune enteropathy triggered by gluten and related prolamines in genetically predisposed individuals. Although CD is a polygenic disease, there is a strong association with genes of the human leukocyte antigen (HLA) region. Most patients present the HLA-DQ2 heterodimer, specifically the DQ2.5 isoform, which is present in around 90-96% of patients of European ancestry. The remaining patients mainly present the HLA-DQ8 heterodimer. This fact and the functional role of these receptors in CD pathogenesis lead us to believe that the presence of these heterodimers is necessary but not enough to develop CD. This confers a very high negative predictive value to the genetic study, almost 100%, although with a low specificity 1,2.
HLA-DQA1 and HLA-DQB1 genes encode the α andβ subunits, respectively, of the heterodimeric protein HLA-DQ or DQ. Genotyping of these loci or the identification of the specific alleles encoding DQ2.5 and DQ8 (HLA-DQA1*05, HLA-DQB1*02, HLA-DQA1*03 and HLA-DQB1*03:02) define the basis of the genetic test used for CD diagnosis.
With the increase in the understanding of CD, HLA studies have gained relevance as a diagnostic tool. This was included in the new CD diagnostic criteria for children and adolescents proposed by the European Society for Paediatric Gastroenterology, Hepatology and Nutrition (ESPGHAN) in 2012 2. The genetic study in adult guidelines has a variable inclusion and relevance, but overall, it is used to support a diagnosis in certain cases. Due to the increased use of genetic studies in CD diagnosis, there is some relevant information that must be considered in order to report and interpret HLA genetic findings. This need has raised some concerns among the members of the Sociedad Española de Enfermedad Celíaca (SEEC), who wanted to address this issue from the working group of Genetics and Immunology. With the aim to establish the recommendations that are referred to in this document, it is important to highlight that the HLA genetic study can be used for a lifelong exclusion of CD, with the consequential social and quality of life implications 3.
Recommendations have been grouped as follows: necessary, advisable, to be avoided and optional information, in a HLA genetic study. In addition, some information aimed to clarify some notions and an example of a genetic report have been included.
NECESSARY INFORMATION
As we have mentioned, the vast majority of CD subjects have the HLA-DQ2 heterodimer, meaning the DQ2.5 isoform and/or the HLA-DQ8 heterodimer. However, there are some patients that carry only one of the alleles encoding DQ2.5. This observation was first described in some European populations 4 and was later ratified in Spain 5, as well as in populations from other geographical regions 6,7,8,9. Therefore, we consider it mandatory that a genetic report includes the information with regard to the following three points:
Reporting whether the subject has the HLA-DQ2 heterodimer, meaning DQ2.5 (presence of HLA-DQA1*05 and HLA-DQB1*02 alleles) and/or HLA-DQ8 (presence of HLA-DQA1*03 and HLA-DQB1*03:02 alleles).
In the absence of HLA-DQ2 (DQ2.5) and HLA-DQ8, information about the presence of one of the alleles encoding DQ2.5 (HLA-DQA1*05 or HLA-DQB1*02) must be included.
Information about how to interpret genetic data must be added, indicating whether the observed genetics is compatible or non-compatible with CD development. With regard to compatible genetics, the presence of HLA-DQ2 (DQ2.5) and/or HLA-DQ8 must be considered and also the presence of only the HLA-DQA1*05 or the HLA-DQB1*02 allele. The fact that the high negative predictive value of the genetic study is present in the absence of the DQ8 haplotype and of both alleles encoding DQ2 should be highlighted 10.
ADVISABLE INFORMATION
The presence of DQ2.5 and DQ8 confers susceptibility to develop CD, but there is also a gene dosage effect. Thus, DQ2.5 individuals have a higher risk of CD when they carry two HLA-DQB1*02 alleles. DQ8 subjects have a higher risk when they are DQ8 homozygous 4. On the other hand, it is advisable to add the information about genotype when full HLA-DQ genotyping is performed. This allows for the detection of possible errors when reporting HLA heterodimers. However, it must also be noted that there are still unknown CD risk alleles. Full genotyping could provide relevant information in the future for physicians or researchers and also for patients. This can be summarized in the following two bullet points, which are highly recommended for inclusion in the genetic report:
1Indicating the genetic dose (one or two copies) of the HLA-DQB1*02 allele and the DQ8 haplotype.
Indicating the full genotype of HLA-DQA1 and HLA-DQB1 loci, i.e., the two specific alleles present in each gene.
Interpretation of the genetic information is not always obvious. It is common to consider a positive result as indicative of CD at the present moment or in the near future. Therefore, it is important to highlight that the relevance of the genetic test is its high negative predictive value (almost 100%). However, it lacks a high positive predictive value and should not be used as a single tool to diagnose CD, as around 35-40% of the general population have the HLA-DQ2 and/or HLA-DQ8 heterodimers and only around 1% develop CD. We recommend clarifying that HLA genetics included in the genetic report is considered as necessary but not enough for CD development.
INFORMATION TO BE AVOIDED
Related to the aforementioned, assertions that suggest that the presence of HLA-DQ2 and/or HLA-DQ8 are indicative of CD at that moment or in the near future must be avoided. In the same manner, non-compatible genetics does not discard CD with a 100% probability. There are certain CD patients lacking HLA genetic risk, although at very low frequency 4,5.
OPTIONAL INFORMATION
The risk to develop CD varies depending on the HLA-DQ genotype present. The risk level of each particular subject can be indicated according to this. When included, we recommend using the following classification:
Very high risk: the presence of HLA-DQ2 (i.e., DQ2.5) with two copies of the HLA-DQB1*02 allele (DQ2.5/DQ2.5, DQ2.5/DQ2.2).
High risk: the presence of HLA-DQ2 (i.e., DQ2.5) with one copy of the HLA-DQB1*02 allele or homozygous for HLA-DQ8 (DQ2.2/DQ7.5 (= DQ2.5 in trans configuration), DQ2.5/DQ8, DQ2.5/DQ7.5, DQ2.5/other, DQ8/DQ8).
Moderate risk: the presence of the HLA-DQ8 and/or the HLA-DQB1*02 allele (DQ8/DQ2.2, DQ2.2/DQ2.2, DQ8/DQ7.5, DQ8/other, DQ2.2/other).
Low risk: the presence of the HLA-DQA1*05: DQ7.5/DQ7.5, DQ7.5/other allele.
No risk alleles.
"Other" indicates another DQ that is different from DQ2.5, DQ8, DQ2.2 and DQ7.5.
Although this classification can be modified, we recommend to avoid the use of "low risk" in HLA-DQ8 individuals or in those carrying only the HLA-DQB1*02 allele. "This can be misunderstood and to consider that it is very unlikely that those individuals present CD. Some differences in the CD risk can be present depending on the specific genotype in the high and moderate risk groups. It is important to highlight that the presence of only the HLA-DQA1*05 allele does not increase CD risk, as it is present at a higher frequency in the general population. However, it cannot be used to discard CD. HLA-DQA1*05 must be considered as compatible with a CD diagnosis.
ADDITIONAL INFORMATION
Table 1 shows the alleles in the HLA-DQA1 and HLA-DQB1 genes and the haplotypes that they form, which are responsible for the different CD-associated DQ heterodimers or isoforms. DQ heterodimers and haplotypes can have a similar nomenclature. However, it should be noted that each subject has two haplotypes, each one inherited from one progenitor. Consequently, the DQ2.5 heterodimer can be present in the absence of the DQ2.5 haplotype. The DQ2.5 heterodimer is encoded by the HLA-DQA1*05 and HLA-DQB1*02 alleles and therefore can appear in the presence of the DQ2.5 haplotype (cis configuration, inherited by only one progenitor); as well as in presence of the DQ2.2 and DQ7.5 haplotypes (trans configuration, each haplotype inherited from one progenitor). It must be considered that the presence of only the HLA-DQA1*03 allele (i.e., in absence of HLA-DQB1*03:02) does not confer a risk to develop CD.
Table 1 HLA-DQ alleles and haplotypes conferring a risk for celiac disease, including their encoded proteins
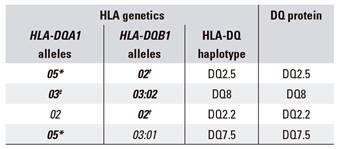
The alleles conferring a risk for celiac disease are in bold. The alleles in plain text are included as they form haplotypes with accompanying risk allele. *It includes HLA-DQA1*05:01 and HLA-DQA1*05:05 alleles. †It includes HLA-DQB1*02:01 and HLA-DQB1*02:02 alleles. ‡It includes HLA-DQA1*03:01 and HLA-DQA1*03:02 alleles.
There are several techniques to identify HLA risk alleles. Some offer the full HLA-DQ genotyping and others only identify the risk alleles. It can be useful to indicate the exact methodology performed in order to know if the information provided in the genetic report is complete.
CONCLUSIONS
CD is a chronic disease with a lifelong treatment: a gluten free diet. This highlights the relevance of a proper diagnosis. The genetic test provides strong support for a diagnosis in certain cases. However, when interpreting or reporting HLA genetic findings, it is necessary to clearly understand several ideas to avoid misdiagnosis.
COMMENTS
Members of the Sociedad Española de Enfermedad Celíaca (SEEC) collaborating in the present report:
Eduardo Arranz. Mucosal Immunology Group. IBGM. Universidad de Valladolid-CSIC. Valladolid (Spain).
José Ramón Bilbao. Department of Genetics, Physical Anthropology and Animal Physiology, Universidad del País Vasco (UPV-EHU), Instituto de Investigación del Hospital Universitario de Cruces. BioCruces. Leioa, Vizcaya (Spain).
Fernando Fernández-Bañares. Service of Gastroenterology. Hospital Universitari Mutua Terrassa. Terrassa, Barcelona (Spain); CIBERehd of Hepatic and Digestive Diseases. Terrassa, Barcelona (Spain).
Juana Jiménez. Service of Clinical Analysis and Biochemistry. Hospital Universitario Severo Ochoa. Leganés, Madrid (Spain).
Teresa Perucho. Laboratory of Genetics. Genyca Innova. Majadahonda, Madrid (Spain).
Eva Ruiz-Casares. Laboratory of Genetics. Genyca Innova. Majadahonda, Madrid (Spain).
Félix Sánchez-Valverde. Pediatric Gastroenterology and Nutrition Unit. Pediatrics Service. Complejo Hospitalario de Navarra. Pamplona, Navarra (Spain).
Juan I. Serrano-Vela. Research and Training. Asociación de Celíacos y Sensibles al Gluten. Madrid (Spain).
BIBLIOGRAFÍA
1. Hadithi M, Von Blomberg BM, Crusius JB, et al. Accuracy of serologic tests and HLA-DQ typing for diagnosing celiac disease. Ann Intern Med 2007;147:294-302. DOI: 10.7326/0003-4819-147-5-200709040-00003 [ Links ]
2. Husby S, Koletzko S, Korponay-Szabo IR, et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J Pediatr Gastroenterol Nutr 2012;54:136-60. DOI: 10.1097/MPG.0b013e31821a23d0 [ Links ]
3. Rodríguez Almagro J, Hernández Martínez A, Lucendo AJ, et al. Health-related quality of life and determinant factors in celiac disease. A population-based analysis of adult patients in Spain. Rev Esp Enferm Dig 2016;108:181-9. DOI: 10.17235/reed.2016.4094/2015 [ Links ]
4. Karell K, Louka AS, Moodie SJ, et al. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol 2003;64:469-77. DOI: 10.1016/S0198-8859(03)00027-2 [ Links ]
5. Fernández-Banares F, Arau B, Dieli-Crimi R, et al. Systematic review and meta-analysis show 3% of patients with celiac disease in Spain to be negative for HLA-DQ2.5 and HLA-DQ8. Clin Gastroenterol Hepatol 2017;15:594-6. [ Links ]
6. Mubarak A, Spierings E, Wolters V, et al. Human leukocyte antigen DQ2.2 and celiac disease. J Pediatr Gastroenterol Nutr 2013;56:428-30. DOI: 10.1097/MPG.0b013e31827913f9 [ Links ]
7. Kotze LM, Nisihara R, Utiyama SR, et al. Absence of HLA-DQ2 and HLA-DQ8 does not exclude celiac disease in Brazilian patients. Rev Esp Enferm Dig 2014;106:561-2. [ Links ]
8. Pallav K, Kabbani T, Tariq S, et al. Clinical utility of celiac disease-associated HLA testing. Dig Dis Sci 2014;59:2199-206. DOI: 10.1007/s10620-014-3143-1 [ Links ]
9. Araya M, Oyarzun A, Lucero Y, et al. DQ2, DQ7 and DQ8 distribution and clinical manifestations in celiac cases and their first-degree relatives. Nutrients 2015;7:4955-65. DOI: 10.3390/nu7064955 [ Links ]
10. Díaz-Redondo A, Miranda-Bautista J, García-Lledo J, et al. The potential usefulness of human leukocyte antigen typing for celiac disease screening: a systematic review and meta-analysis. Rev Esp Enferm Dig 2015;107:423-9. DOI: 10.17235/reed.2015.3758/2015 [ Links ]
Received: October 04, 2017; Accepted: February 01, 2018











 text in
text in 


