My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
International Microbiology
Print version ISSN 1139-6709
INT. MICROBIOL. vol.7 n.1 Mar. 2004
| RESEARCH ARTICLE | ||||
| Competition for polymers | |||
| Summary. Three heterotrophic bacterial strains, isolated from organic particles of the upper water column of the Equatorial Atlantic, taken during a cruise on the R/V METEOR (1997), were investigated concerning their physiological and phylogenetic properties using classic microbiological and modern molecular-biological methods. All isolates are gram-negative rods able to use polymers such as cellulose, chitin or starch as sole carbon source. The phylogeny of these isolates was investigated by fluorescence in situ hybridization (FISH) and 16S rDNA sequencing. The three isolated strains belong to the Cytophaga/Flavobacteria, γ-Proteobacteria (Marinobacter sp.), and α-Proteobacteria (Sulfitobacter pontiacus). In order to study succession during growth on polymers naturally occurring in marine habitats, FISH was used as a new approach to detect cells from different phylogenetic clusters in the course of a single growth experiment. Mixed cultures consisting of the isolated strains in equal amounts were incubated with cellulose, chitin or starch. Isolate 4301-10/2, a member of the γ-Proteobacteria, dominated in mixed cultures growing on cellulose, chitin, or starch after only 10 days, with 55, 60, and 95%, respectively, of cells hybridizing with 4´,6-diamidino-2-phenylindole (DAPI). [Int Microbiol 2004; 7(1): 13–18, 2004] Key words: FISH · particle-attached bacteria · marine bacteria · mixed culture · substrate utilization | |||
| Competición por polímeros entre bacterias heterótrofas aisladas de partículas en el Atlántico ecuatorial Resumen. Tres cepas de bacterias heterótrofas, aisladas de partículas orgánicas de la parte superior de la columna de agua del Atlántico ecuatorial, tomadas durante un crucero con el R/V Meteor (1997), fueron investigadas respecto a sus propiedades fisiológicas y filogenéticas usando tanto técnicas microbiológicas clásicas como métodos moleculares modernos. Todas las cepas aisladas son bacilos Gram-negativos con capacidad de usar polímeros como celulosa, quitina o almidón como única fuente de carbono. La filogenia de estas cepas se investigó mediante hibridación in situ por fluorescencia (FISH) y secuenciación del rDNA 16S. Las tres cepas aisladas pertenecen a las Cytophaga/Flavobacterias, las γ-Proteobacterias (Marinobacter sp.), y las α-Proteobacterias (Sulfitobacter pontiacus). Para estudiar la sucesión a lo largo del crecimiento con polímeros presentes de forma natural en ambientes marinos, se usó FISH para detectar las células de diferentes grupos filogenéticos a lo largo de un experimento de crecimiento. Cultivos mixtos consistentes en las cepas aisladas en cantidades iguales se incubaron con celulosa, quitina o almidón. Transcurridos sólo 10 días, la cepa 4301-10/2, que es una γ-Proteobacteria, dominó los cultivos mixtos con celulosa, quitina o almidón, con el 55, 60 y 95%, respectivamente, de las células hibridadas con 4´,6-diamidino-2-fenilindol (DAPI). [Int Microbiol 2004; 7(1):13-18] Palabras clave: FISH · bacterias adheridas a partículas · bacterias marinas · cultivo mixto · utilización de substratos | Competição pelos polímeros entre bactérias heterotróficas isoladas de partículas do Atlântico equatorial Palavras chave: FISH · bactérias aderidas as partículas · bactérias marinhas · cultivo mixto · utilização de substratos |
Introduction
A crucial mechanism in the global cycle of carbon is the biologically mediated export of particulate organic carbon from the ocean's surface to the deep sea. Marine bacteria greatly influence the transport processes of organic carbon in the water column [5]. Free-living bacteria, most of which have been classified as α-Proteobacteria or γ-Proteobacteria, exist in marine habitats, but bacteria attached to marine particles play a more important role [1,9] because the dominant fraction-involved in the transport of biogenic carbon from the surface water to the deep-sea bottom-consists of macroscopic aggregates, such as marine snow [2]. In comparison to the surrounding water, aggregates have higher concentrations of nutrients and show elevated microbial activities [6]. They provide enriched microenvironments of organic matter in the oligotrophic ocean and are thus hotspots of microbial respiration, which cause a fast and efficient respiratory turnover of particulate organic carbon in the sea [5,6]. Marine snow in the mesopelagic water column originates from aggregating phytoplankton cells, fecal pellets, or mucus from zooplankton [6,12,20]. Smaller particles, which are composed of polysaccharides, transparent exopolymer particles (TEP) or proteins, are two to three orders of magnitude more abundant than larger particles [3,16]. TEP could serve as substrates and microhabitats that provide attached bacteria with physical refuges from predators unable to feed on surfaces [3]. Bacteria inhabiting sinking particles are often bigger and, on a cellular basis, more active than free-living ones [13,30] and have been classified as Cytophaga/Flavobacteria, Planctomyces, α-or γ-Proteobacteria [1,9,22]. Only small quantities of hydrolysates are taken up by these bacteria attached to particles [28]. The excess of dissolved organic matter forms elongated "plumes," streaming behind sinking particles, leading to population explosions of free-living bacteria [14]. However, inhibition mechanisms are also widespread among marine bacteria. Antagonistic interactions among aggregate-colonizing bacteria and protists-as well as interacting bacteria-have a large, mutual influence on the composition of an aggregate-colonizing bacterial population. The formation of acylated homoserine lactones (AHL) leads to different strategies to exert predator pressure or growth competition with other bacteria [17,19].
It was the aim of this study to examine an artificially rebuilt consortium composed of bacterial strains, isolated from sinking particles of the Equatorial Atlantic. Three bacterial strains, representing three different phylogenetic groups, were isolated from particles in order to study their microbial decomposition potential with special emphasis on the degradation of polymers. As a new application for the distinction of different phylogenetic groups regarding their competition behavior during growth on polymeric substrates, fluorescence in situ hybridization (FISH) was used.
Materials and methods
Origin and cultivation of heterotrophic bacterial strains. During a cruise in the Equatorial Atlantic with R/V METEOR in February/March 1997, water samples were taken from a depth of 50 m using 10-l Niskin-bottle water samplers (Hydrobios, Kiel, Germany). The material was filtrated through 10-µm pore size Isopore membranes (Millipore), which were then transferred into sterile seawater. Dilution series were made with aggregate-containing water from station no. 4301-10 near Gran Canaria (29º 09.2´ N; 15º 30.3´ W). Agar plates with ASNIII medium (basic medium), modified and prepared according to Rippka et al. [23], contained 0.1% yeast extract, 3.4% NaCl, 0.2% MgCl2·6H2O, 0.05% KCl, 0.075% NaNO3, 0.35% MgSO4·7H2O, 0.05% CaCl2·2H2O, 0.002% Na2CO3, 0.001% K2HPO4·3H2O, 0.00015% Fe-NH4-citrate, 0.000001% vitamin B12, 0.05% actidione, and 1.8% agar. The plates were then inoculated with material from the highest dilution rate in order to isolate dominant species. Cultures were incubated at 27ºC, and single colonies were transferred into fresh medium to obtain pure cultures.
Utilization of substrates. Growth of three isolates on amylopectin, chitin, chitobiose, glycogen, maltose, and melibiose was examined using agar plates with ASNIII medium without yeast extract and Fe-NH4-citrate, but with 0.2% (w/v) of the above-mentioned carbon substances. Growth of the isolates on acetate, N-acetyl-D-glucosamine, L-alanine, cellobiose, citrate, fructose, glucose, mannose, and sucrose (5 mM each) was examined utilizing liquid ASNIII medium without yeast extract and Fe-NH4-citrate by following the increase of the optical density at 600 nm. Enzymatic activities of cellulase, gelatinase, amylase, lecithinase and DNase were tested by streaking the isolates on artificial seawater agar plates (ASNIII with 0.1% w/v yeast extract, supplemented with either 1% w/v cellulose, 0.4% w/v gelatin, 2% w/v starch, 5% w/v egg-yolk emulsion, or 0.2% w/v DNA and 0.005% w/v methylgreen, respectively). Cellulase activity of the isolates was tested by streaking them onto agar plates with ASNIII and cellulose (1% w/v) as sole carbon source. The plates were incubated at 27ºC for 4-6 days. Cellulose hydrolysis was proved by the development of colonies on the agar plates, while lecithinase production was detected by the appearance on egg-yolk agar plates of cloudy precipitations around colonies (modified after [26]). Starch hydrolysis was documented using Lugol iodine solution to demonstrate colorless areas around bacterial growth, and gelatin degradation by using picric acid to visualize unhydrolyzed polymers (modified after [29]). DNA hydrolysis was analyzed by treating the plates with 2 M HCl to reveal undigested DNA (modified after [27]).
Determination of G + C content. DNA was isolated with Qiagen Genomic-tips 100/G (Qiagen) as described in the Qiagen Genomic DNA Handbook [21]. The DNA was purified by ethanol precipitation and collected in 0.1× standard saline citrate (SSC). For further purification, the DNA was desalted using a Microcon Centrifugal Filter Devices (Amicon) following the manufacturer's instructions. The melting profiles were determined using a spectrophotometer (Beckman) equipped with a high-performance temperature controller. Solutions containing ~15 µg DNA in 325 µl 0.1× SSC were heated from 40 to 95ºC by 0.1ºC min-1; optical density was recorded at 260 nm every 10 s. The G+C content of the DNA was calculated according to the following equation, reported by Marmur and Doty [18]:
mol% G + C=([Tmsample+{90.5-TmE. coli}] -69.3)/0.41
The melting point (Tm) was taken as the mid-point between the upper and lower asymptotes.
FISH. Formaldehyde-fixed (7% final concentration) cells from a single colony of each isolate were transferred onto Teflon-coated microscope slides and immobilized by drying at 46ºC. After dehydration and fixation with 50, 80, and 96% (v/v) ethanol, slide-fixed cells were hybridized with oligonucleotide probes EUB338 (5´-GCT GCC TCC CGT AGG AGT-3´), ARCH915 (5´-GTG CTC CCC CGC CAA TTC CT-3´), CF319a (5´-TGG TCC GTG TCT CAG TAC-3´), ALF968 (5´-GGT AAG GTT CTG CGC GTT-3´), BET42a (5´-GCC TTC CCA CTT CGT TT-3´), and GAM42a (5´-GCC TTC CCA CAT CGT TT-3´) (modified after [10]). All probes were CY3-labeled and synthesized by Interactiva (Ulm, Germany). The cells were counterstained with DAPI (1 µg/ml) and incubated in the dark for 5 min. After washing (70% ethanol, bidistilled water) and air drying, the slides were mounted with a 5:1 (v/v) mixture of Citifluor AF1 (Citifluor, London, UK) and Vectashield (Linaris, Wertheim, Germany). The cells were examined using an epifluorescence microscope (Zeiss Axiolab) with filter sets for DAPI (filter set 02, Zeiss, Germany) and CY3 (Chroma HQ 41007).
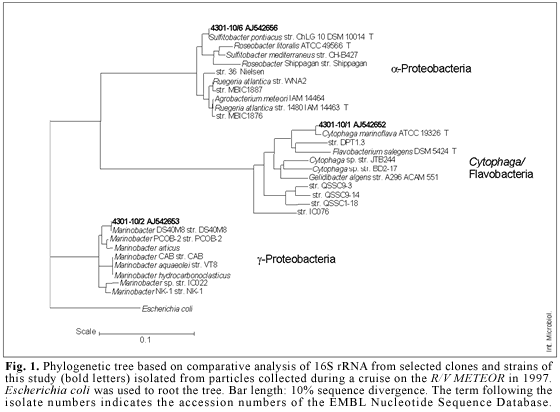
Phylogenetic analysis. Genomic DNA (isolation as described above) was amplified using REDTaq (Sigma Aldrich) and the bacterial primers 10-30F (5´-AGA GTT TGA TCM TGG CTC GA-3´) and 1542R (5´-AGA AAG GAG GTG ATC CAR CC-3´). The amplification mixture consisted of 25-30 ng template DNA, 0.4 µM of each primer, 200 µM dNTP-Mix, 4 µl bovine serum albumin (2 mg/ml), 2 mM MgCl2, 5 µl reaction buffer (100 mM Tris-HCl, 500 mM KCl, 11 mM MgCl2, and 0.1% gelatin), 2 U REDTaq polymerase, and sterile water to a total volume of 50 µl. PCR amplification was confirmed by gel electrophoresis on a 1% agarose gel stained with ethidium bromide. PCR products were purified with QIAquick Gel Extraction Kits (Qiagen) and desalted by using 0.025-µm nitrocellulose filters (Millipore). Sequencing was conducted with the bacterial primers 10-30F (see above) and 518F (5´-CCA GCA GCC GCG GTA AT-3´) by MWG Biotech, Germany. Sequence data were analyzed with the PHYLIP software package [http://rdp.cme.msu.edu/cgis/phylip.cgi]. A phylogenetic tree was reconstructed using maximum-likelihood and neighbor-joining analyses. The 16S rDNA sequences from isolates generated in this study were deposited in the EMBL Nucleotide Sequence Database under the accession numbers AJ542652, AJ542653, and AJ542656.
Degradation of polymers. In order to prepare mixed, triplicate cultures with equal amounts of cells for competition experiments, samples of 1-4×106 cells/ml were taken from cultures pre-grown on the various polymers in basic medium without yeast extract and Fe-NH4-citrate at 27ºC on a rotary shaker (125 rpm) for up to 48 days. Cellulose or chitin served as carbon source for the three isolates 4301-10/1, 4301-10/2, and 4301-10/6, while starch was used only for the first two. Aliquots of 1.4 ml were removed with an Eppendorf pipette from the growing cultures at different time intervals in order to analyze the bacterial composition of the mixed culture by the FISH method. These samples were fixed with 100 µl sterile-filtrated formaldehyde (37% v/v) and incubated for 5 min at room temperature. Then, the mixture was filtrated through 0.2-µm Isopore membranes (47 mm diameter) and rinsed with 1 ml sterile bidistilled water. The filters were stored at -20ºC until hybridization. After thawing, filter sections approximately one-eighth in size were cut out with a sterile scalpel and mounted onto sterile slides. Hybridization with oligonucleotide probes EUB338, CF319a, ALF968, GAM42a, counterstaining with DAPI, and mounting for microscopy were done as described above.
Results and Discussion
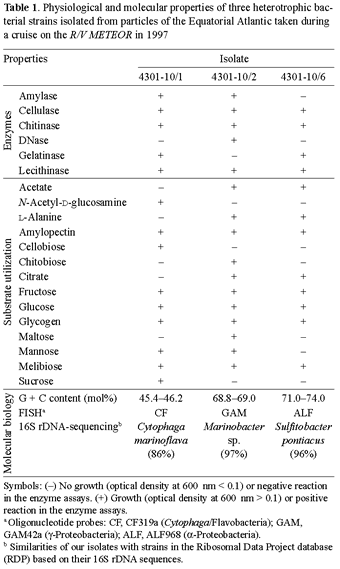
Physiological and phylogenetic properties of the isolates. The three isolates obtained were examined with regard to their enzymatic machinery, their ability to use different substances as sole carbon source, and their molecular-biological properties (Table 1). The isolates differed strongly in their content of extracellular enzymes such as amylase, DNase, and gelatinase. All three strains were able to produce cellulase and chitinase. Likewise, the isolates varied in their ability to metabolize different low-molecular-weight carbon compounds (mono-or dimers), but they were able to utilize most of the substances provided. Strain 4301-10/2 used 11 out of 14 organic compounds tested, while the other two strains metabolized only nine or eight substrates, respectively. Particle-associated bacteria often have an increased metabolic activity compared to that of free-living bacteria, which are adapted to a largely oligotrophic environment [4]. Different molecular-biological properties were examined to taxonomically characterize the three isolates of this study. Table 1 shows that the G + C content of isolate 4301-10/1 was very low, 45.4-46.2 mol%, which is typical for Cytophaga/Flavobacteria [11], while that of isolates 4301-10/2 and 4301-10/6 was higher, 68.8-69.0 mol% and 71.0-74.0 mol%, respectively. The investigated strains hybridized with probes specific for members of Cytophaga/Flavobacteria (4301-10/1), γ-Proteobacteria (4301-10/2), or α-Proteobacteria (4301-10/6). These results are in accordance with those obtained by 16S rDNA sequencing. By comparison with the Ribosomal Database Project II [8], the closest relative of isolate 4301-10/1 is Cytophaga marinoflava, with a similarity of 86%. Isolates 4301-10/2 and 4301-10/6 showed higher similarities to Marinobacter sp. (97%) and Sulfitobacter pontiacus (96%).
Figure 1 shows a phylogenetic tree including the three investigated isolates. All isolated strains are typical representatives of marine habitats. The most abundant phylogenetic types detected in macroaggregate-associated bacterial populations from investigations in the Santa Barbara Channel fell within the Cytophaga/Flavobacteria group [9]. Representatives of this group produce exoenzymes and are able to degrade a wide spectrum of polymeric compounds, such as proteins, polysaccharides, chitin, and nucleic acids [9,24]. Acinas and co-workers [1] found divergent distributions of phylogenetic groups on marine particles. The most abundant species in offshore Western Mediterranean waters belonged to the γ-Proteobacteria. This discrepancy could be explained by the very different nature of macroaggregates, which consist of large amounts of detrital organic matter. The genus Marinobacter is known for its absolute requirement of sodium ions [11]. Members of the γ-Proteobacteria appear to be involved in the very early stages of colonization of surfaces. In the α-Proteobacteria cluster, the genus Roseobacter, which is closely related to the genus Sulfitobacter, is known for rapidly colonizing surfaces, too. Members of α-Proteobacteria could be found free-living as well as particle-attached [1,9].
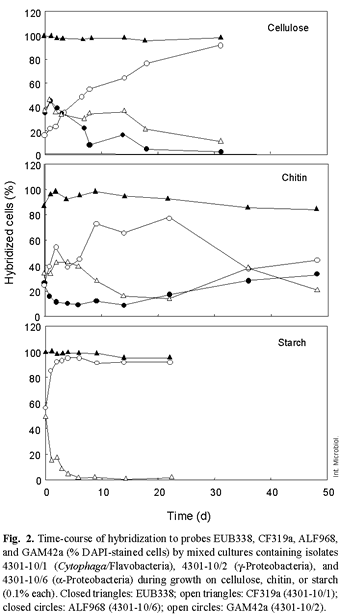
Degradation of polymers. All three isolates were able to hydrolyze cellulose and chitin, but only isolates 4301-10/1 and 4301-10/2 degraded starch additionally. Therefore, three mixed cultures with either cellulose, chitin or starch as sole carbon source were incubated for 22-48 days in order to study their ability to compete for polymers as growth substances (Fig. 2). In order to distinguish the strains from each other and to quantify them during the experiment, samples were withdrawn at different times during growth and analyzed by FISH.
Hybridization with oligonucleotide probes EUB338 (Bacteria), CF319a (Cytophaga/Flavobacteria), ALF968 (α-Proteobacteria), and GAM42a (γ-Proteobacteria) allowed visualization of the cells for counting. Counterstaining with DAPI was used to record the total amount of cells in each sample. EUB338-hybridized cells comprised 90-100% of total cell counts for all three assays. Due to the low concentration of isolate 4301-10/2 at the beginning of the experiment with cellulose, isolates 4301-10/1 and 4301-10/6 dominated the mixed culture during the first 3 days of incubation. Subsequently, isolate 4301-10/2 continued to increase, reaching 90% of the cell population after 31 days, whereas the other two isolates decreased to 10 and 2%, respectively.
In contrast to the experiment with cellulose, all isolates showed nearly the same amount of cells (30 or 50% of total cell counts) at the beginning of the growth experiments with chitin or starch. Irrespective of the carbon source offered, isolate 4301-10/2 dominated the mixed cultures, reaching 80% and more than 90% of the total amount of cells in the chitin-, respectively, starch-containing medium following 9 (chitin) or 2 (starch) days of incubation. Nevertheless, the growth behavior of all the isolates differed while growing on chitin or starch (see middle and lower panels of Fig. 2). When growing on chitin, the amounts of cells of isolates 4301-10/1 and 4301-10/6 decreased to 20% and 5% respectively during the first 14 days of incubation. During this time, isolate 4301-10/2 started to decrease slowly before increasing to about 55%, whereas the cell counts of isolate 4301-10/1 increased up to 44% (day 36 of incubation) before decreasing to less than 20%. The growth behavior of isolate 4301-10/6 differed from that of the other two isolates; the amount of cells of this isolate was reduced to 5% after 2 days of incubation, remained constant for about 10 days and then slowly increased up to 30%. By contrast, isolate 4301-10/2 reached more than 90% after 3 days and outcompeted isolate 4301-10/1 to less than 10% of the total cell amount in the culture containing starch. Isolate 4301-10/2 (γ-Proteobacteria) dominated a consortium with isolates 4301-10/1 (Cytophaga/ Flavobacteria) and 4301-10/6 (α-Proteobacteria) in all cases when growing on cellulose, chitin, or starch. Acinas and co-workers [1] stated that assemblages of large cells, belonging mostly to particle-attached γ-Proteobacteria, grow rapidly and override other microbial groups.
In the decomposition of marine phytoplankton, changes in population structure correlate with the ability to degrade high-molecular-weight organic components [28]. Laboratory investigations on the long-term microbial degradation of chitin have shown a decrease in the amount of attached bacteria and an increase in the number of free-living bacteria [15]. Under optimal laboratory conditions, in which only one nutrient is growth-limiting, it could be expected that one organism totally outcompetes all of the others in the culture if its efficiency of substrate utilization is better [7]. In addition to "overgrowing" competitors, another way to compete against concurring bacteria inhabiting the same niche is the development of bacterium-bacterium antagonistic interactions, such as the production of inhibitory substances, as shown by Long and Azam [17]. These authors observed that heterotrophic marine bacteria produce inhibitory substances against other marine bacteria. Members of the γ-Proteobacteria (Alteromonadales and Vibrionales) were the most prolific producers of such inhibitory compounds. Both possibilities, better enzymatic machinery and/or the production of inhibitory substances, could explain the strong dominance of isolate 4301-10/2, belonging to the γ-Proteobacteria. There are many interacting factors that influence the development of microbial consortia, and the competition for substrates is a major evolutionary driving force in the microbial world. A much more precise knowledge of these processes occurring on the highly diverse particles in the ocean will be important for understanding the complexity of global carbon-flux pathways. As shown in this study, FISH is a valuable scientific tool that cane be used in growth experiments to distinguish bacteria from different phylogenetic groups competing for a sole substrate.
Acknowledgements. We are grateful to the officers and crew of the R/V METEOR, as well as B. Karwath and N. Zatloukal for their competent assistance in the retrieval of water samples. We also wish to extend our sincere thanks to Kerstin Probst for assisting in proofreading the English. This research project was funded by the Deutsche Forschungsgemeinschaft (Sonderforschungsbereich 261, Teilprojekt A1, Bremen University, contribution no. 359).
References
1. Acinas SG, Antón J, Rodríguez-Valera F (1999) Diversity of free-living and attached bacteria on offshore western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol 65:514-522 [ Links ]
2. Alldredge AL, Silver MW (1988) Characteristics, dynamics and significance of marine snow. Prog Oceanogr 20:41-82 [ Links ]
3. Alldredge AL, Passow U, Logan BE (1993) The abundance and significance of a class of large, transparent organic particles in the ocean. Deep-Sea Res 40:1131-1140 [ Links ]
4. Ayo B, Unanue M, Azua I, Gorsky G (2001) Kinetics of glucose and amino acid uptake by attached and free-living marine bacteria in oligotrophic waters. Mar Biol 138:1071-1076 [ Links ]
5. Azam F (1998) Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696 [ Links ]
6. Caron DA, Davis PG, Madin LP, Sieburth JMcN (1982) Heterotrophic bacteria and bacterivorous protozoa in oceanic macroaggregates. Science 218:795-797 [ Links ]
7. Christensen BB, Haagensen JAJ, Heydorn A, Molin S (2002) Metabolic commensalism and competition in a two-species microbial consortium. Appl Environ Microbiol 68:2495-2502 [ Links ]
8. Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442-443 [ Links ]
9. DeLong EF, Franks DG, Alldredge AL (1993) Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38:924-934 [ Links ]
10. Eilers H, Pernthaler J, Glöckner FO, Amann R (2000) Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol 66:3044-3051 [ Links ]
11. Holt JG, Krieg NR, Sneath PHA, Staley JT, Williams ST (1994) Bergey's Manual of Determinative Bacteriology. Williams and Wilkins, Baltimore, pp 86, 483-514 [ Links ]
12. Iriberri J, Herndl GJ (1995) Formation and microbial utilization of amorphous aggregates in the sea: ecological significance. Microbiologia SEM 11:309-322 [ Links ]
13. Iriberri J, Unanue M, Barcina I, Egea L (1987) Seasonal variation in population density and heterotrophic activity of attached and free-living bacteria in coastal waters. Appl Environ Microbiol 53:2308-2314 [ Links ]
14. Kiørboe T, Jackson GA (2001) Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol Oceanogr 46:1309-1318 [ Links ]
15. Kirchner M (1995) Microbial colonization of copepod body surfaces and chitin degradation in the sea. Helgoländer Meeresuntersuchungen 49:201-212 [ Links ]
16. Long RA, Azam F (1996) Abundant protein-containing particles in the sea. Aquat Microb Ecol 10:213-221 [ Links ]
17. Long RA, Azam F (2001) Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol 67:4975-4983 [ Links ]
18. Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109-118 [ Links ]
19. Pernthaler J, Posch T, Simek K, Vrba J, Amann R, Psenner R (1997) Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol 63:596-601 [ Links ]
20. Pilskaln CH, Honjo S (1987) The fecal pellet fraction of biogeochemical particle flux to the deep sea. Global Biogeochem Cycles 1:31-48 [ Links ]
21. Qiagen GmbH (1997) Qiagen Genomic DNA Handbook for blood, cultured cells, tissue, mouse tails, yeast, bacteria (gram- and some gram+). Qiagen GmbH, Hilden, Germany
22. Rath J, Wu KY, Herndl GJ, DeLong EF (1998) High phylogenetic diversity in a marine-snow-associated bacterial assemblage. Aquat Microb Ecol 14:261-269 [ Links ]
23. Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY (1979) Generic assignment, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol 111:1-61 [ Links ]
24. Reichenbach H (1989) Genus I. Cytophaga. In: Staley JT, Bryant MP, Pfennig N, Holt J (eds) Bergey's Manual of Systematic Bacteriology, vol 3. Williams and Wilkins, Baltimore, pp 2015-2050 [ Links ]
25. Simon M, Alldredge AL, Azam F (1990) Bacterial carbon dynamics on marine snow. Mar Ecol Prog Ser 65:205-211 [ Links ]
26. Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for General and Molecular Bacteriology. American Society for Microbiology, Washington DC, pp 614-622 [ Links ]
27. Smith PB, Hancock GA, Rhoden DL (1969) Improved methods for detecting deoxyribonuclease production by bacteria. Appl Microbiol 18:991-993 [ Links ]
28. Smith DC, Simon M, Alldredge AL, Azam F (1992) Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139-142 [ Links ]
29. Süßmuth R, Eberspächer J, Haag R, Springer W (1987) Biochemisch-Mikrobiologisches Praktikum (in German). Thieme, Stuttgart, pp 73-74 [ Links ]
30. Turley CM, Stutt ED (2000) Depth related cell specific bacterial leucine incorporation rates on particles and its biogeochemical significance in the NW Mediterranean. Limnol Oceanogr 45:419-425 [ Links ]