My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
International Microbiology
Print version ISSN 1139-6709
INT. MICROBIOL. vol.7 n.1 Mar. 2004
| RESEARCH ARTICLE | |||
|
| |||
| Nucleotide sequence and expression of the ncr nickel and cobalt resistance in Hafnia alvei 5-5
| ||
| Summary. The structural genes for the nickel and cobalt resistance of the conjugative plasmid pEJH 501 of Hafnia alvei 5-5, contained on a SalI-EcoRI fragment of 4.8 kb, were cloned and sequenced. The DNA sequence included five genes in the following order: ncrA, ncrB, ncrC, ncrY, and ncrX. The predicted amino acid sequences of ncrA were homologous to the amino acid sequences of nreB of Achromobacter xylosoxidans 31A. Expression of ncr with the T7 RNA polymerase-promoter system allowed Escherichia coli BL21 (DE3) to overexpress NcrA, NcrB, and NcrC but not NcrY, and NcrX. The apparent molecular masses of NcrA, NcrB, and NcrC were 30, 33, and 17 kDa, respectively. Primer-extension analysis showed that ncr mRNA started at nucleotide position 23 upstream from ncrA. The promoter region of the ncr operon possessed a strong, putative -35 element of ð32-type promoter sequence, and transcriptional 'lacZ fusion studies indicated that the -35 element influenced ð32-specific transcription. [Int Microbiol 2004; 7(1): 27-34, 2004] Key words: plasmid pEJH501 · operon NcrABCYX · ð32-type promoter sequence · nickel-resistant bacteria | |||
| |||
| Secuencia nucleotídica y expresión del determinante ncr de resistencia a níquel y cobalto en Hafnia alvei 5-5 Resumen. Los genes estructurales de la resistencia a níquel y cobalto del plásmido conjugativo pEJH 501 de Hafnia alvei 5-5, contenido en un fragmento SalI-EcoRI de 4,8 kb, fueron clonados y secuenciados. La secuencia de DNA incluye cinco genes en el siguiente orden: ncrA, ncrB, ncrC, ncrY, y ncrX. Las secuencias de aminoácidos equivalentes a ncrA fueron homólogas a las secuencias de aminoácidos codificadas por nreB en Achromobacter xylosoxidans 31A. La expresión de los genes ncr mediante el sistema promotor de la RNA polimerasa T7 permite a Escherichia coli BL21 (DE3) sobreexpresar NcrA, NcrB, y NcrC, pero no NcrY ni NcrX. Los pesos moleculares aparentes de NcrA, NcrB y NcrC fueron 30, 33, y 17 kDa, respectivamente. El análisis de extensión de los cebadores mostró que el mRNA de ncr se iniciaba a una distancia de 23 nucleótidos corriente arriba del ncrA. La región promotora del operón ncr posee una fuerte secuencia promotora de tipo ð32 en la posición -35, y estudios transcripcionales de fusión con ´lacZ indicaron que el elemento situado en -35 influye sobre la transcripción específica de ð32. [Int Microbiol 2004; 7(1):27-34] Palabras clave: plásmido pEJH501 · operón NcrABCYX · secuencia promotora de tipo ð32 · bacterias resistentes a níquel | Sequência nucleotídica e expressão do determinante ncr de resistência à niquel e cobalto em Hafnia alvei 5-5 Resumo. Os genes estruturais de resistência à níquel e cobalto do plasmídeo conjugativo pEJH 501 de Hafnia alvei 5-5, contido em um fragmento SalI-EcoRI de 4800 pares de bases (pb), foram clonados e sequenciados. A sequência de DNA inclue cinco genes na seguinte ordem: ncrA, ncrB, ncrC, ncrY, e ncrX. As sequências de aminoácidos equivalentes à ncrA foram homólogas às sequências de aminoácidos codificadas para nreB em Achromobacter xylosoxidans 31A. A expressão dos genes de ncr mediante o sistema promotor da RNA polimerase T7 permite a Escherichia coli BL21 (DE3) supra-expressar NcrA, NcrB, e NcrC, porém não os genes NcrY e NcrX. Os pesos moleculares aparentes de NcrA, NcrB e NcrC foram 30, 33, e 17 kDa, respectivamente. A análise de extensão de iniciadores mostrou que o mRNA de ncr era iniciado a uma distância de 23 nucleotídeos antes de ncrA. A região promotora do operon de ncr possue uma sequência promotora putativa forte do tipo ð32 na posição -35, e estudos transcricionais de fusão com ´lacZ indicaram que o elemento situado na posição -35 tem influência na transcrição específica ð32. [Int Microbiol 2004; 7(1):27-34] Palavras chave: plasmídeo pEJH501 · operon NcrABCYX · sequência promotora do tipo ð32 · bactérias resistentes ao níquel |
Introduction
Nickel is a trace element that serves as an essential constituent of enzymes, such as hydrogenase, CO dehydrogenase, and urease in bacteria [10]. However, excess nickel ions interact with cellular components, such as amino acids and nucleotides, resulting in a disturbance of enzyme activity, DNA replication, transcription, and translation [1,16]. Plasmid-mediated resistance to nickel has been described in Ralstonia metallidurans CH34 [7,8,15,17,32-34] and Achromobacter xylosoxidans 31A [28-30]. Resistance is due to the action of an inducible, operon-encoded energy-dependent specific efflux system that secretes the cation from the cell thereby lowering the intracellular concentration of the toxic compound [31].
The cnr (cobalt-nickel resistance) operon of the R. metallidurans CH34 plasmid pMOL28 consists of cnrYXHCBA. The genes cnrCBA encode a membrane-bound protein complex catalyzing an energy-dependent efflux of cobalt and nickel [5]. The mechanism of action of the CnrCBA complex may be that of a proton/cation antiporter based on the considerable similarity of CnrCBA amino acid sequences to those of CzcCBA (for cadmium-zinc-cobalt resistance). The topological orientation and function in the membrane of CzcCBA have been well-studied in strain CH34 [20,21,23]. The Cnr regulatory genes cnrYXH are arranged in a region upstream of the structural genes and are responsible for full transcription of the CnrCBA structural resistance genes. CnrH, a 21-kDa protein, belongs to a sigma factor of the extracytoplasmic function (ECF) family, whose members share a helix-turn-helix motif at the carboxy terminus [18,24,37]. CnrX, which may function as a periplasmic sensor, contains histidine residues that probably bind nickel ions. CnrY is a trans-acting regulatory protein thought to act as a repressor or anti-sigma factor [7].
The ncc (nickel-cobalt-cadmium resistance) operon of the A. xylosoxidans 31A pTOM9 comprises seven open reading frames (ORFs), nccYXHCBAN, and the encoded proteins share strong homology to proteins encoded by cnrYXHCBA [30]. The NccCBA complex also shows close similarities to the CzcCBA complex, which seems to be a three-component cation-proton antiporter [30]. NccH probably belongs to a family of ECF ð70-like proteins, as it has the conserved regions of the ECF sigma factors. NccX contains several histidine residues and seems to be capable of binding nickel ions; therefore, the protein may function as a periplasmic sensor. NccY is a trans-acting regulatory protein and down-regulates the operon [30].
In addition to the ncc locus, A. xylosoxidans 31A contains another nickel resistance locus, nre, on plasmid pTOM9, which confers a low level of resistance [8,30]. Gene nreB is induced by nickel and may be involved in resistance by efflux coupled to a chemiosmotic gradient. The topological orientation of this gradient in the membrane has been elucidated largely by comparison with proteins of the major facilitator superfamily (MFS) transporters [8].
In this work, we have determined the complete nucleotide sequence of the 4.8-kb SalI-EcoRI fragment containing the ncr (nickel-cobalt resistance) operon from the 70-kb plasmid pEJH501 of Hafnia alvei 5-5. Five ORFS for the genes ncrABCYX were found. The products of the predicted translation product of ncrA and ncrB were consistent with those of nreB and nreA [22].
Materials and methods
Bacterial strains, plasmids, and growth conditions. The bacterial strains and plasmids used in this study are described in Table 1. Growth conditions for Hafnia alvei 5-5 were described previously [22]. The bacterial culture was deposited in the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) in Braunschweig, Germany, with the number DSM 15533. Escherichia coli strains were grown at 37ºC in Tris mineral medium supplemented with 0.3% (w/v) gluconate as carbon source [17]. Amino acids, when necessary, were added at a concentration 20 µg/ml after filtration. For maintenance of plasmid markers, filter-sterilized solutions of antibiotics were added, as appropriate, to the following final concentrations: ampicillin, 100 µg/ml, kanamycin 30 µg/ml, and chloramphenicol 15 µg/ml for E. coli. Clones of E. coli DH5α harboring recombinant pUC plasmids were identified on LB agar plates containing 100 µg ampicillin/ml, 40 µg 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside/ml, and 0.2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Duchepa, Netherlands).
Chemicals, reagents, and enzymes. Analytical grade NiCl2·6H2O (Sigma, St. Louis, MI, USA) was prepared as 1.0 M stock solution and sterilized by autoclaving. All enzymes were purchased from Promega (Madison, WI, USA), TakaRa (Japan), and New England Biolab (Beverly, MA, USA). [γ-32P] ATP (1000 Ci/mmol) and [α-35S] dATP (1000 Ci/mmol) were purchased from Amersham (UK). β-Galactosidase activity was measured as published before [19] and is reported in Miller units.
Genetic techniques. Standard molecular genetic techniques were as described by Sambrook et al. [25] or as per the manufacturer's instructions unless mentioned specially. DNA sequences were obtained by the dideoxy-mediated termination method of Sanger using an ABISEQ automatic sequencer. Nucleotide and amino acid sequences were analyzed with the sequence analysis software DNAMAN (Lynnon Biosoft), Clone Manager 5 (Scientific and Educational Software), and Align Plus (Scientific and Educational Software). Homology searches were done using the BLAST search algorithms located at the National Center for Biotechnology Information site (http://www.ncbi.nlm.nih.gov/) on the World Wide Web. The sequence has been submitted to GenBank (accession no. AF322866). Hydropathy profiles were determined using the method of Hopp and Woods [12,14].
Transcript analysis. Total RNA was isolated from uninduced cells or cells exposed to 300 µM NiCl2 for 2 h at 37ºC during growth in Tris mineral medium by using the RNeasy total RNA preparation kit (Qiagen, Santa Clara, Calif., USA). The transcription start site was identified by primer extension using primer 5´-ACGCGTCGACTGCAGGAATTCACACTTTTAATC-3´, which is complementary to nucleotides +28 to +49 downstream of P1, respectively. The primer was end-labeled using T4 polynucleotide kinase (Promega, Madison, WI, USA) and [γ-32P] ATP following standard protocols. For primer-extension experiments, a modification of the Promega protocol was used. Two pmol of the primers were incubated with 30 µg RNA in 10 µl hybridization buffer (50 mM KCl, 25 mM Tris-HCl, pH 8.3) at 65-75ºC for 2 min and allowed to cool. AMV RT (1 U) was used to extend the primer in a reaction mixture containing 50 mM KCl, 10 mM MgCl2, 10 mM dithiothreitol (DTT), 0.5 mM spermidine, and 1 mM deoxynucleotide triphosphate. The reaction was carried out in a total volume of 20 µl at 42ºC for 30 min. After phenol-chloroform extraction, the extended product was precipitated by the addition of 0.1 volume of 3 M sodium acetate and three volumes of ethanol. The pellet was washed with 75% ethanol, dried, and dissolved in 10 µl H2O. The unlabeled primer was used to generate a nucleotide sequence ladder using a Sequenase version 2.0 DNA sequencing kit (Amersham Life Science, Cleveland, Ohio, USA) with [35S] dATP. Primer-extension products were separated in an 8 M urea/6% polyacrylamide gel in parallel with the sequencing reactions in order to map the transcription initiation site.
Protein expression. The ncr genes were expressed by the T7 RNA polymerase-promoter system [36]. The genes were cloned into a T7-promoter-containing vector (pT7-6), and the resulting plasmid was transformed into E. coli BL21 (DE3) bearing the T7 RNA polymerase gene (λ DE3 lisogen) for expression of target proteins. Cells bearing plasmid were grown at 37ºC overnight in Luria Bertani (LB) medium containing 50 µg ampicillin/ml). The culture was diluted 100-fold into 5 ml fresh LB medium, containing ampicillin alone or ampicillin and nickel (250 µM), and was grown at 37ºC. When the culture reached the mid-exponential phase, 1.0 mM IPTG was added to induce gene expression. Cultivation was continued for an additional 2 h. Cells were pelleted by centrifugation and suspended in 500 µl of 10 mM Tris-HCl buffer (pH 7.0). Cells were then frozen at -20ºC for 16 h and disrupted on ice using an ultrasonicator (Lab Line, USA) at a continuous setting output of 10 times for 10 s. After centrifugation at 9,000 rpm at 4ºC for 20 min, the supernatant was added to the same volume of 10 mM Tris-HCl buffer (pH 7.0) and the pellet was dissolved in 20 µl 2× Tricine sample buffer [0.08 M Tris-Cl/SDS (pH 6.8), 24% (v/v) glycerol, 8% (w/v) SDS, 0.2 M DTT, 0.02% (w/v) Coomassie blue G-250]. Proteins were analyzed on a 10% Tris-Tricine/SDS-polyacrylamide gel [30% acrylamide/0.8% bisacrylamide, Tris×HCl/SDS (pH 8.45), glycerol, 10% (w/v) ammonium persulfate, TEMED].
Results
Cloning of determinant for nickel and cobalt resistance (ncr). A 4.8-kb SalI-EcoRI DNA fragment was cloned in several steps from the 70-kb plasmid pEJH501 of H. alvei 5-5 into the SalI-EcoRI sites of phagemid vector pBluescript II KS (+), forming plasmid pHF14 [22]. When plasmid pHF14 was transformed into E. coli, the cells grew in the presence of 10 mM nickel chloride. However, plasmids containing subcloned DNA fragments conferred only weak resistance to nickel, comparable to those with the plasmid pHF14 (Fig. 1).
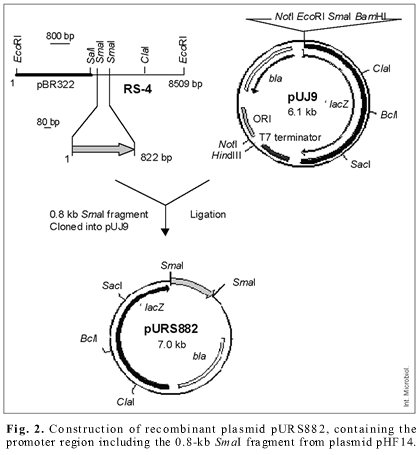
These results indicated that the 4.8-kb SalI-EcoRI DNA fragment contains the intact determinant of nickel resistance and functions in E. coli. To determine whether a promoter was derived from sequences on the parent plasmid pEJH501, the 822-bp SmaI fragment from pHF14 was cloned into the SmaI site of plasmid pUJ9 containing the promoterless reporter gene 'lacZ (Fig. 2). Cells of E. coli DH5a carrying the resulting 'lacZ fusion plasmid pURS882 expressed enhanced β-galactosidase activity, as the concentration of nickel increased, while showing low levels in the absence of inducer. These results indicated that this fragment contains the promoter sequence of the intact nickel resistance determinant of pEJH501, whose expression is induced by nickel chloride.
Nucleotide sequence of the ncr operon. The locus for nickel and cobalt resistance was localized to the 4.8-kb SalI-EcoRI fragment of DNA (Fig. 1). The complete nucleotide sequence of the cloned fragment was determined in both strands by sequence walking. Five potential open reading frames (ORFs) for protein-coding regions were identified by computer analysis along with the proposed initiation codons, stop codons, the proposed ribosome-binding sites, and the deduced amino acid sequences. These protein-coding regions were oriented in the same direction, and each ORF contained an AUG translation start codon together with a properly spaced ribosome-binding site (the nucleotide sequence of the 4800-bp DNA fragment of nickel resistance determinants is deposited under the accession no. AF322866).
The first ORF, ncrA, starts 403 nucleotides from the beginning of the insert and extends for 831 nucleotides, corresponding to 277 amino acids. ncrA has a weak ribosome-binding site, AGC, located ten nucleotides upstream from the initiation ATG codon. The ncrA product is a homologue of NreB from A. xylosoxydans 31A and of NrsD from Synechocystis sp. strain PCC6803 [6,13]. Both the amino acid composition (77% nonpolar amino acids) and the hydropathy profile (data not shown) indicate that the nreA gene product should be localized primarily in the membrane.
The second reading frame, ncrB, starting at position 1243 and ending at position 2145, encodes a protein of 301 amino acids that is a homologue of NreA of A. xylosoxydans 31A [15]. The overall sequence identity between NreA and NcrB was 78 % at the amino acid level. The presumed ribosome-binding site of ncrB, GG, is located 6 nucleotides upstream from the initiation ATG codon.
The third reading frame, ncrC, beginning at position 2260 and ending at position 2715, comprises 456 bp, corresponding to 152 amino acids. The presumed ribosome-binding site, GGA, is located 15 nucleotides upstream from the initiation ATG codon. There was no similarity to known proteins involved in heavy metal resistance. There were no intergenic sequences between ncrA and ncrB, and no promoter-like sequence was found upstream from ncrB and ncrC, which suggested that ncrABC, conferring nickel resistance, should be read as a single transcript.
The fourth reading frame, ncrY, beginning at position 3709 and ending at position 3897, encodes 63 amino acids. The presumed ribosome-binding site, GGA, is located 8 nucleotides upstream from the initiation ATG codon. RT-PCR was used to search for the transcript of ncrY in E. coli DH5a after cloning into plasmid pHF14. Total RNA was isolated under induced condition and used as the template for RT reactions, using the primer at the 3´ end of ncrY. Using cDNA obtained from RT with ncrY primer, the PCR product was amplified for ncrY. In addition, the putative promoter was identified in the DNA region preceding ncrY and highly conserved between cnrYp, cnrCp, nccYp, and nccCp promoters. Previous work showed that transposon insertion in this region caused E. coli to constitutively express resistance [22]. This established that ncrY is expressed and may function as a trans-acting regulator.
The fifth reading frame, ncrX, starting at position 4354 and ending at position 4434, encodes a protein of 27 amino acids. Previous work showed that transposon insertions in this region also caused the constitutive expression of resistance [22]. Those data also suggested that this gene functions as trans-acting regulator.
>RT-PCR experiments were done to search for a transcript corresponding to the region between ncrC and ncrY. There was no PCR product when a primer complementary to the region flanking the 3´ end of ncrY and the 5´ end of ncrC was used (data not shown). These data indicated that ncrABC and ncrY were separated by 993 nucleotides that were not transcribed.
Primer extension mapping of the ncrABC promoter. To determine the location of the ncrABC promoter, the transcriptional start site was mapped by primer extension (Fig. 3). One distinct extension product was obtained, corresponding to the A residue at a position 28 nucleotides upstream from the initiation ATG codon. The deduced start site is therefore 5´-C361CCCCGCCAGGATAATGCTTGTCATTTTTTT-3´ (the +1 nucleotide A375 is underlined). As shown in Fig. 3, the -10 region (CCCCCGCCA) upstream of the transcriptional start is separated from the -35 region (TATCAGGCAACTA) by 14 nucleotides. This is a match to the canonical sequence recognized by ð32-RNA polymerase.
Expression of ncrABC in E. coli. To determine whether ncrABC was transcribed from the intrinsic promoter and then expressed, the 4.8-kb SalI-EcoRI fragment was inserted into the expression vector pT7-6, which yielded pTF714, in which the ncr operon was placed under the control of the T7 promoter. E. coli BL21 (DE3), harboring pNRS148, was grown in the presence of inducer, sonicated, and then fractionated into membrane and soluble parts. The inducible proteins were present in large amounts in the membrane fractions and were determined to have apparent molecular masses of 30, 33 and 17 kDa, as predicted for NcrA, NcrB, and NcrC (Fig. 4).
Discussion
The cnr system enables R. metallidurans CH34 to grow in the presence of 3 mM nickel and 5 mM cobalt ions [31]. The ncc system encodes high-level resistance to nickel (40 mM), cobalt (20 mM), and cadmium (1 mM) in A. xylosoxidans 31A [29]. The nre system also confers the ability to tolerate nickel (3 mM) to A. xylosoxidans 31A [30]. Hafnia alvei 5-5 was isolated from a soil-litter mixture underneath the canopy of the nickel-hyperaccumulating tree Sebertia acuminata in New Caledonia. The bacterium is able to grow in the presence of 30 mM nickel as well as 2 mM cobalt [27,35]. The level of nickel resistance is similar to that produced by ncc genes and higher than resistances conferred by the cnr and nre systems. It was of interest, therefore, to identify the components of the nickel pump in H. alvei 5-5 and to compare them with those of the cnr, ncc, and nre systems. As a first step, the genes were sequenced and the amino acid sequences of their potential protein products were deduced. The DNA sequence of a 4.8-kb SalI-EcoRI fragment contained five ncr genes, ncrA, ncrB, ncrC, ncrX, and ncrY. The ncrA product is a homologue of NreB from A. xylosoxydans 31A [15] and NrsD from Synechocystis sp. strain PCC6803 [6,13]. Previous work suggested that NreB was responsible for low-level nickel resistance by efflux and closely related to MFS transporters [6]. From the hydropathic profile of the NcrA protein, there were 15 regions of 18 or more amino acid residues in length with a hydropathy index greater than 1.5, which is indicative of possible membrane-spanning α-helices for a transporter. NcrA has a broad range of metal-ion substrates, i.e. nickel, cobalt and zinc, while NreB is more specific for nickel [22].
Although no significant homology was found between NcrB, NcrC and proteins listed in the database, the overall hydropathy profile is similar to that of membrane proteins. TnPhoA´-1 mutagenesis has shown that the NcrB and NcrC are necessary for nickel resistance and transport [22]. This implies that NcrB and NcrC may in some way modify the substrate specificity and activity catalyzed by NcrA. Examples of proteins that change the substrate specificity of enzymes include ArsC. This protein changes the substrate specificity catalyzed by ArsA and ArsB thereby allowing recognition of both arsenate and arsenite [3] and ð factors, which alters the recognition site of RNA polymerase during carbon starvation and heat-shock response [9,11]. Based on these data, we propose a model in which NcrA, NcrB, and NcrC form a membrane-bound complex catalyzing cobalt and nickel efflux.
The promoter region of nreABC was characterized by primer extension. The major reverse transcript corresponded to a start site that was preceded by a ð32-recognized sequence, indicating that Eð32 would bind to this site with high affinity. Our results support this possibility in that transcriptional fusion of lacZ to this promoter produced high levels of β-galactosidase to heat shock response. This was the first instance in which a gene concerned with nickel resistance has been shown to be regulated by a ð32 promoter.
The level of nickel resistance by NcrABC was higher than that produced by NreA. One possibility is that the NcrABC complex provides a more effective mechanism of nickel resistance than NreA. This mechanism has been observed for the ars operon of the R-factor R773 from E. coli [2]. Cells expressing arsB exhibited an intermediate level of arsenite resistance compared with cells expressing both arsA and arsB. Arsenite exclusion by ArsB was coupled to electrochemical energy, while transport by the ArsA-ArsB complex was coupled to ATP hydrolysis. Another possibility is that the NcrABC complex is highly expressed, at levels comparable to those producing a higher level of resistance. The promoter region of nreABC was shown to be regulated by ð32, which also directs core RNA polymerase to transcribe heat-shock promoters. The transient increase in expression of heat-shock genes after temperature up-shift results from increased transcription initiation at heat-shock promoters, which is mediated by a transient 20-fold increase in the amount of ð32 per cell [9]. Higher expression of NcrABC would be a more effective mechanism of providing resistance to nickel ions present in heavily contaminated environments, resulting in a selective pressure for its evolution.
Acknowledgements. The authors wish to acknowledge the financial support of the Korea Research Foundation made in the program year of 1998 (015-D00216). We are also thankful for the financial support of the Alexander von Humboldt-Stiftung, Germany.
References
1. Brintzinger H (1963) The structures of adenosine triphosphate-metal ion complexes in aqueous solution. Biochem Biophys Acta 77:343-345 [ Links ]
2. Carlin A, Shi W, Dey S, Rosen BP (1995) The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol 177:981-986 [ Links ]
3. Chen CM, Mobley HLT, Rosen BP (1985) Separate resistance to arsenate and arsenite (antimonite) encoded by the arsenical resistance operon of R-factor R773. J Bacteriol 161:758-763 [ Links ]
4. De Lorenzo V, Herrero M, Timmis KN (1990) Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol 172:6568-6572 [ Links ]
5. Dong Q, Mergeay M (1994) Czc/Cnr efflux: a three-component chemiosmotic antiport pathway with a 12-transmembrane-helix protein. Mol Microbiol 14:185-187 [ Links ]
6. Garcia-Dominguez M, Lopez-Maury L, Florencio FJ, Reyes JC (2000) A gene cluster involved in metal homeostasis in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 182:1507-1514 [ Links ]
7. Grass G, Grobe C, Nies DH (2000) Regulation of the cnr cobalt and nickel resistance determinant from Ralstonia sp. Strain CH34. J Bacteriol 182:1390-1398 [ Links ]
8. Grass G, Fan B, Rosen BP, Lemke K, Schlegel HG, Rensing C (2001) NreB from Achromobacter xylosoxidans 31A is a nickel-induced transporter conferring nickel resistance. J Bacteriol 183:2803-2807 [ Links ]
9. Gross CA (1996) Function and regulation of the heat shock proteins. In: Neidhardt FC, Curtiss R, Lin EC (eds) Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, pp 1382-1399 [ Links ]
10. Hausinger RP (1987) Nickel utilization by microorganisms. Microbiol Rev 51:22-42 [ Links ]
11. Hengge-Aronis R (1993) Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in Escherichia coli. Cell 72:165-168 [ Links ]
12. Hopp TP, Woods KR (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA 78:3824-3828 [ Links ]
13. Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miuajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S (1996) I. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3:109-136 [ Links ]
14. Kyte J, Doolittle RF (1982) A Simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105-132 [ Links ]
15. Liesegang H, Lemke K, Siddiqui RA, Schlegel HG (1993) Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J Bacteriol 175:767-778 [ Links ]
16. Martin RB (1988) Nickel ion binding to amino acids and peptides. In: Sigel H, Sigel A (eds) Metal Ions in Biological Systems, vol. 23. Nickel and its Role in Biology. Marcel Dekker, New York, pp 123-164 [ Links ]
17. Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, van Gijsegem F (1985) Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol 162: 328-334 [ Links ]
18. Missiakas D, Raina S (1998) The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol 28:1059-1066 [ Links ]
19. Nies DH (1992) CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol 174:8102-8110 [ Links ]
20. Nies DH (1995) The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J Bacteriol 177:2707-2712 [ Links ]
21. Nies DH, Silver S (1995) Ion efflux systems involved in bacterial metal resistances. J Ind Microbiol 14:186-199 [ Links ]
22. Park JE, Young, KE, Schlegel HG, Rhie HG, Lee HS (2003) Conjugative plasmid mediated inducible nickel resistance in Hafnia alvei 5-5. Int Microbiol 6:57-64 [ Links ]
23. Rensing C, Pribyl T, Nies DH (1997) New functions for the three subunits of the CzcCBA cation-proton antiporter. J Bacteriol 179:6871-6879 [ Links ]
24. Rouviere PE, deLasPenas A, Mecsas J, Lu CZ, Rudd KE, Gross CA (1995) RpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J 14:1032-1042 [ Links ]
25. Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York [ Links ]
26. Sancar A, Hack M, Rupp WD (1979) Simple method for identification of plasmids coded proteins. J Bacteriol 137:692-693 [ Links ]
27. Schlegel HG, Cosson J-P, Baker AJM (1991) Nickel-hyperaccumulating plants provide a niche for nickel-resistant bacteria. Bot Acta 104:18-25 [ Links ]
28. Schmidt T, Schlegel HG (1989) Nickel and cobalt resistance of various bacteria isolated from soil and highly polluted domestic and industrial wastes. FEMS Microbiol Ecol 62:315-328 [ Links ]
29. Schmidt T, Stoppel RD, Schlegel HG (1991) High-level nickel resistance in Alcaligenes xylosoxydans 31A and Alcaligenes eutrophus KTO2. Appl Environ Microbiol 57:3301-3309 [ Links ]
30. Schmidt T, Schlegel HG (1994) Combined nickel-cobalt-cadmium resistance encoded by the ncc locus of Alcaligenes xylosoxidans 31A. J Bacteriol 176:7045-7054 [ Links ]
31. Sensfuss C, Schlegel HG (1988) Plasmid pMOL28-encoded resistance to nickel is due to specific efflux. FEMS Microbiol Lett 55:295-298 [ Links ]
32. Siddiqui RA, Schlegel HG (1987) Plasmid pMOL28-mediated inducible nickel resistance in Alcaligenes eutrophus strain CH34. FEMS Microbiol Lett 43:9-13 [ Links ]
33. Siddiqui RA, Schlegel HG, Meyer M (1988) Inducible and constitutive expression of pMOL28-encoded nickel resistance in Alcaligenes eutrophus N9A. J Bacteriol 170:4188-4193 [ Links ]
34. Siddiqui RA, Benthin K, Schlegel HG (1989) Cloning of pMOL28-encoded nickel resistance genes and expression of the genes in Alcaligenes eutrophus and Pseudomonas spp. J Bacteriol 171:5071-5078 [ Links ]
35. Stoppel RD, Schlegel HG (1995) Nickel-resistant bacteria from anthropogenically nickel-polluted and naturally nickel-percolated ecosystems. Appl Environ Microbiol 61:2276-2285 [ Links ]
36. Tabor S, Richardson CC (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA 82:1074-1078 [ Links ]
37. Tibazarwa C, Wuertz S, Mergeay M, Wyns L, van der Lelie D (2000) Regulation of the cnr cobalt and nickel resistance determinant of Ralstonia eutropha (Alcaligenes eutrophus) CH34. J Bacteriol 182:1399-1409 [ Links ]