Meu SciELO
Serviços Personalizados
Journal
Artigo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Acessos
Acessos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares em
SciELO
Similares em
SciELO -
 Similares em Google
Similares em Google
Compartilhar
International Microbiology
versão impressa ISSN 1139-6709
INT. MICROBIOL. vol.7 no.2 Jun. 2004
| RESEARCH ARTICLE | |||
|
| |||
| Protein and glycoprotein content of lymphocystis disease virus (LCDV)
Summary. The polypeptide and glycoprotein composition of eight strains of the fish-pathogenic lymphocystis disease virus (LCDV) isolated from gilt-head seabream (Sparus aurata), blackspot seabream (Pagellus bogaraveo), and sole (Solea senegalensis) were determined. The protein electrophoretic patterns of all LCDV isolates were quite similar regardless of the host fish, showing two major proteins (79.9 and 55.6 kDa) and a variable number of minor proteins. Three groups of LCDV isolates were distinguished according to the number and molecular masses of the minor proteins. Eight glycoproteins were detected inside viral particles of LCDV 2, LCDV 3 and LCDV 5 isolates, but only seven glycoproteins were found inside viral particles of LCDV 1, LCDV 4, LCDV 6, LCDV 7, and LCDV 11 isolates and the reference virus ATCC VR 342 by using five lectins. LCDV glycoproteins were mainly composed of mannose and sialic acid. These glycoproteins could be part of an external viral envelope probably derived from the host cell membrane. [Int Microbiol 2004; 7(2):121-126] Key words: lymphocystis virus · lectins · tumor lesions in fish | ||
| |||
Contenido proteico y glicoproteico del virus de la linfocistis (LCDV) Resumen. En el presente trabajo se ha determinado la composición polipeptídica y se caracterizaron las glicoproteínas de ocho cepas del virus de la linfocistis (LCDV), un patógeno de peces, aisladas de dorada (Sparus aurata), besugo (Pagellus bogaraveo) y lenguado (Solea senegalensis). Todos los aislados de LCDV presentaban patrones electroforéticos similares, pero no idénticos, obtenidos por coloración azul brillante de Coomassie y nitrato de plata, independientemente de la especie hospedadora. Los patrones electroforéticos mostraban dos proteínas mayoritarias de una masa molecular estimada de 79,7 y 55,6 kDa y un número variable de proteínas minoritarias. Se clasificaron los aislados en tres grupos a partir de las diferencias en el número total de bandas polipeptídicas y en el perfil de proteínas minoritarias. Se detectaron ocho glicoproteínas en los viriones de los aislados LCDV 2, LCDV 3 y LCDV 5, pero sólo siete glicoproteínas en los viriones de los aislados LCDV 1, LCDV 4, LCDV 6, LCDV 7 y LCDV 11 y en el virus de referencia ATCC VR 342, usando cinco lectinas. Las glicoproteínas de LCDV están fundamentalmente compuestas por manosa y ácido siálico, y pueden formar parte de la envuelta externa viral que deriva, probablemente, de la membrana de la célula hospedadora. [Int Microbiol 2004; 7(2):121-126] Palabras clave: virus de la linfocistis · lectinas · lesiones tumorales en peces | Conteúdo proteico e glicoproteico do vírus da linfocístis (LCDV) Resumo. Foi determinada a composição em polipéptidos e a caracterização das glicoproteínas de oito isolados de vírus da linfocístis (LCDV), um patógeno de peixes, isolado de dourada (Sparus aurata), besugo (Pagellus bogaraveo) e de linguado (Solea senegalensis). Todos os isolados LCDV apresentaram padrões electroforéticos semelhantes, mas não idênticos, obtidos por coloração azul brilhante de Coomassie e nitrato de prata, indiferentemente da espécie hospedeira. Ambos apresentaram duas bandas polipeptídicas principais, cujos pesos moleculares estimados são 79,7 e 55,6 kDa. Podem ser observadas, entre os isolados LCDV, diferenças ligeiras no número total de bandas polipeptídicas e no padrão proteico inferior, o que nos permite classificá-las em três grupos. Foram detectadas oito glicoproteínas em partículas virais dos isolados LCDV 2, LCDV 3 e LCDV 5, mas foram somente detectadas 7 glicoproteínas nas partículas virais LCDV 1, LCDV 4, LCDV 6, LCDV 7 e LCDV 11 e no vírus de referência ATCC VR 342, usando cinco lectinas. Os resultados obtidos indicam que as glicoproteínas LCDV são compostas principalmente por manose e ácido siálico, e poderão fazer parte de um envelope viral externo, provavelmente, derivado da membrana celular do hospedeiro. [Int Microbiol 2004; 7(2):121-126] Palavras chave: vírus de linfocistis · lectinas · lesões tumorais em peixes |
Introduction
Lymphocystis disease virus (LCDV) is the causative agent of lymphocystis disease, a tumor-like disease of fish characterized by the development of clusters composed of hypertrophied fibroblasts with cytoplasmic inclusions [16]. These cells are individually encapsulated by a hyaline extracellular matrix. Lymphocystis disease affects over 100 different wild and cultured fish species, causing important economic losses. The main host fish for LCDV in the Mediterranean region is gilt-head seabream (Sparus aurata, L.) [12].
LCDV has been included in the genus Lymphocystivirus, belonging to the family Iridoviridae, which comprises four genera: Iridovirus, Chloriridovirus, Ranavirus and Lymphocystivirus [17]. Previous studies have shown that the genera Ranavirus, Iridovirus and Lymphocystivirus include structurally related viruses, since all of them are composed of similar protein units, which contribute to the icosahedral outline structure [9]. Several authors have described the presence of external envelopes in some species of iridovirus, such as frog virus 3 (FV3), derived from a process of budding through the host cell membrane [4]. In addition, a previous biochemical characterization indicated the presence of carbohydrates in the virion, suggesting the existence of glycoproteins [15]. These glycoproteins seem to be responsible for attachment of the virus to cell receptors and are the ideal target for new vaccines. In this study, the polypeptide compositions of different fish LCDV isolates were compared, and the glycoproteins of LCDV virions were characterized.
Materials and methods
Virus isolation and purification. LCDV strains analyzed and compared for protein and glycoprotein composition were isolated from different host species in Spain (Table 1). LCDV strain Leetown NFH (ATCC VR 342) was used as reference strain. Viruses were propagated in SAF-1, a cell line obtained from gilt-head seabream fibroblasts [2]. Cells were cultured and maintained using Leibovitz medium (L-15) (Gibco), supplemented with 1% antibiotic solution (10,000 IU penicillin G/ml, 10 mg streptomycin/ml) (Gibco), 2% L-glutamine (Sigma) and 15% fetal bovine serum (FBS) (LabClinics).
For virus titration, cells were seeded into 24- or 96-well cell culture plates and incubated at 20ºC. The viral dilution infecting 50% of the cell cultures was considered as the end point dilution (TCID50/ml). SAF-1 cells were inoculated with LCDV isolates and, at 15 days post-inoculation, they were harvested, frozen and thawed three times, and sonicated for 20 min in an ultrasonic bath at 40W. This suspension was clarified by centrifugation, and viral particles concentrated by centifugation for 60 min at 60,000 × g at 4ºC. Viruses were resuspended in TNE buffer (0.05 M Tris-HCl, 0.1 M NaCl, 0.001 M EDTA, pH 7.4), layered onto 3-ml gradients of 20-60% (w/w) sucrose, and centrifuged for 2 h at 70,000 × g at 4ºC. The virus band was collected and centrifuged for 1 h at 40,000 × g at 4ºC.
Gel electrophoresis. SDS-PAGE was carried out according to the method of Laemmli [11]. Purified virus concentration was measured using the Bicinchonic Acid Kit for protein determination (Sigma). Five and three mg of viral proteins were detected by Coomassie brilliant blue and silver nitrate staining, respectively. Samples were stacked in 4% acrylamide (15 mA) and separated using 5-15% gradient acrylamide gels (20 mA). Protein molecular mass standards (BioRad) used were: Myosin, 210 kDa; β-galactosidase, 127 kDa; bovine serum albumin, 84 kDa; ovalbumin, 49.5 kDa; carbonic anhydrase, 35.3 kDa; soybean trypsin inhibitor, 28.1 kDa; lysozyme, 20.5 kDa; and aprotinin, 7 kDa. Proteins were stained with Coomassie brilliant blue (Sigma) and silver nitrate (Silver Staining Kit Protein, Amersham Pharmacia Biotech). Gels were preserved by drying.
Glycoprotein detection. In order to detect LCDV glycoproteins, the Glycan Differentiation (Boehringer Mannheim) kit was used, in which the specific binding of lectins to carbohydrates is assayed. The following digoxigenin-labeled lectins (Roche) were used: GNA (Galanthus nivalis agglutinin), which recognizes terminal mannose, α(1-3), α(1-6) or α(1-2) linked to mannose; SNA (Sambucus nigra agglutinin), which recognizes sialic acid linked α(2-6) to galactose; MAA (Maackia amurensis agglutinin), which recognizes sialic acid linked α(2-3) to galactose, and combined with SNA is suitable for identifying both sialyated carbohydrate chains and types of sialic acid linkage; PNA (peanut agglutinin), which recognizes the core disaccharide galactose β(1-3)N-acetylgalactosamine and is thus suitable for identifying O-glycoside-linked carbohydrate chains; and DSA (Datura stramonium agglutinin), which recognizes galactose β(1-4)N-acetylglucosamine in complex and hybrid N-glycans, in O-glycans, and N-acetylglucosamine in O-glycans.
Glycoproteins were transferred onto Immobilon-PVDF membranes (BioRad) after gel electrophoretic separation. The transfer buffer was composed of 25 mM Tris (pH 8.9), 192 mM glycine, 20% methanol and 0.1% SDS. Electroblotting was carried out in a vertical transblot apparatus (TE-42, Hoefer Scientific Instruments) at 75 mA for 18 h. Lectin specificity was demonstrated using the following control glycoproteins: carboxypeptidase Y for GNA; transferrin for SNA; fetuin for SNA, MAA and DSA; and asialofetuin for PNA and DSA. To produce a positive signal, 0.1 µg of the control glycoproteins transferrin, fetuin and carboxypeptidase Y, and 1 µg asialofetuin, are required.
For glycoprotein detection, membranes were treated according to manufacturer´s instructions. Briefly, membranes were blocked for 30 min, washed twice in TBS (0.05 M Tris-HCl, 0.15 M NaCl, pH 7.5) for 10 min, once with TBS supplemented with 1 mM MgCl2, 1 mM MnCl2, 1 mM CaCl2 (pH 7.5), and then incubated for 1 h with 1 mg of each lectin (GNA, SNA, DSA, MAA and PNA)/ml in 50 mM Tris-HCl, 0.05% sodium azide (pH 7.0). Membranes were washed three times with TBS for 10 min and incubated with polyclonal sheep anti-digoxigenin antibody conjugated with alkaline phosphatase (750 U/ml) (1/10 dilution) for 1 h. Excess antibody was washed in TBS three times for 10 min. Membranes were stained with NBT/X-phosphate (4-nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolyl phosphate). In order to stop the reaction, membranes were rinsed with bidistilled water and dried on paper towels.
Results
Infectivity characteristics. Cytopathic effects (CPE) were observed on SAF-1 monolayers after 2-10 days of incubation at 20ºC. The CPE consisted of the rounding and enlargement of infected cells and the formation of cytoplasmic inclusions. The yields of isolated viruses ranged from 1 × 104 TCID50/ml to >1 × 109 TCID50/ml (Table 1).
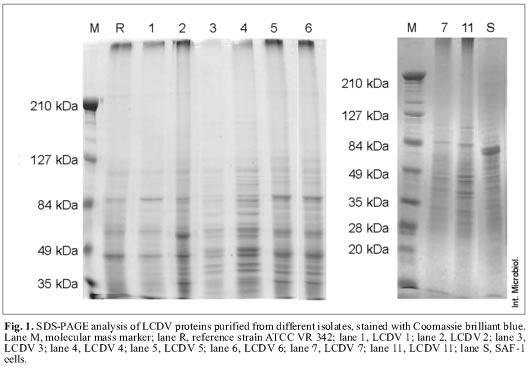
Viral protein patterns. All LCDV isolates showed similar electrophoretic patterns when gels were stained with Coomassie brilliant blue, regardless of the isolation source. These patterns were characterized by two major polypeptides with estimated molecular masses of 79.7 and 55.6 kDa (Fig. 1).
LCDV isolates were classified into three groups according to the number and molecular masses of minor proteins. The reference virus LCDV ATCC VR 342 and the LCDV 1 isolate were included in the first group, characterized by the presence of 14 proteins with estimated molecular masses ranging from 43 to 104.7 kDa. Major protein molecular masses in this group were 44.5, 55.6 and 79.7 kDa. LCDV 2, LCDV 5 and LCDV 6 form the second group, these isolates showed 27 proteins, with estimated molecular masses ranging from 34 to 226 kDa. Major proteins coincided with the above group, although they also included two additional proteins with molecular masses of 37.6 and 33.9 kDa. LCDV 3, LCDV 4, LCDV 7 and LCDV 11 constitute the third group, with a total of 23 proteins. This group shows a clearly different electrophoretic profile compared to the above mentioned groups. Major proteins have molecular masses of 79.7, 76.9, 55.6, 52.8, 46, 43.8, 40.2 and 37.6 kDa.
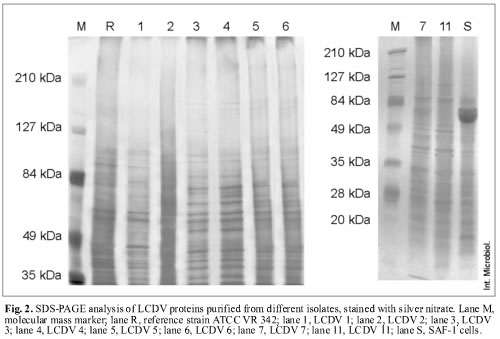
Silver-nitrate staining allowed the specific detection of sialoglycoproteins, and, in addition, the differentiation into three protein patterns (Fig. 2). This technique detected more proteins than Coomassie blue staining. Applying this technique, 21 proteins were found for the reference virus LCDV ATCC VR 342 and LCDV 1, with estimated molecular masses ranging from 30.5 to 136.7 kDa. LCDV 2, LCDV 5 and LCDV 6 isolates showed 30 proteins each, with molecular masses ranging from 29.5 to 187.5 kDa. In addition, LCDV 3, LCDV 4, LCDV 7 and LCDV 11 isolates showed 31 proteins, with molecular masses ranging from 29.7 to 187.6 kDa (Fig. 2).
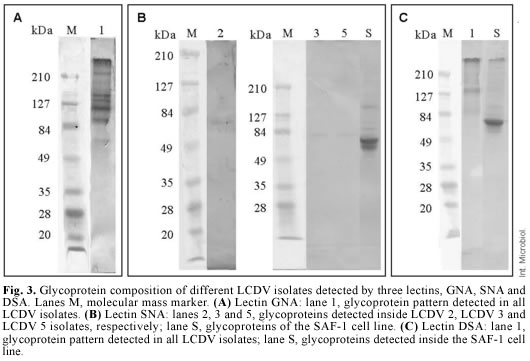
Identification of glycoproteins. The use of five lectins allowed the detection of eight glycoproteins inside the viral particles of LCDV 2, LCDV 3 and LCDV 5 isolates, whereas only seven glycoproteins were detected inside LCDV 1, LCDV 4, LCDV 6, LCDV 7, LCDV 11 isolates and the reference virus ATCC VR 342. The GNA lectin detected six glycoproteins in all the LCDV isolates tested, whose estimated molecular masses were 210, 145, 134, 123, 104 and 77 kDa (Fig. 3). These glycoproteins have N-glycan chains with a high proportion of mannose. A glycoprotein with an estimated molecular mass of 84 kDa was detected by the SNA lectin in LCDV 3 and LCDV 5 (Fig. 3). This lectin also detected a glycoprotein with a different molecular mass (76 kDa) in the LCDV 2 isolate (Fig. 3).
No glycoproteins were detected with the SNA lectin in the remaining LCDV isolates tested. This reaction indicates the presence of sialic acid terminally linked α(2-6) to galactose or N-acetylgalactosamine. The DSA lectin detected one 84-kDa glycoprotein in all LCDV isolates, with N-acetylglucosamine linked to galactose by a β(1-4) linkage (Fig. 3). The MAA lectin identified two glycoproteins in all LCDV isolates, but their molecular masses were similar to those of glycoproteins from non-inoculated SAF-1 cells, used as negative control. The PNA lectin did not identify any glycoprotein, which suggests that none of the LCDV isolates had glycoproteins with N-acetylgalactosamine linked to galactose by a β(1-3) linkage.
Discussion
The present study identified the viral polypeptides of several LCDV isolates and the presence of glycoproteins in purified LCDV particles. Although some differences were found between the electrophoretic patterns of these isolates, the major proteins were similar (79.7 and 55.6 kDa), and wide differences were not observed between the proteins of viral particles isolated from different fish species (gilt-head seabream, sole and blackspot seabream). Flügel et al. [7] studied the protein patterns of LCDV strains isolated from three fish species, flounder (Platichthys flesus), plaice (Pleuronectes platessa) and dab (Limanda limanda), obtaining similar profiles. Although all viral protein profiles had common major polypeptides, with molecular masses of 63 and 50 kDa, there were some differences between certain polypeptide patterns, particularly between LCDVs isolated from flounder and dab.
Polypeptide patterns described in this study differ from those obtained by Kelly et al. [10] in other iridoviruses. Similarities among protein patterns of LCDV virions isolated from different hosts indicate that these profiles do not depend on host species. Previous studies characterizing LCDV showed the presence of a high level of carbohydrates, suggesting the existence of glycoproteins [15]. Of the eight glycoproteins that we detected, six had high levels of mannose, whereas LCDV2, LCDV 3 and LCDV 5 isolates had one glycoprotein containing sialic acid and another glycoprotein containing N-acetylglucosamine linked to galactose.
The presence of glycoproteins is generally associated with enveloped viruses. However, previous studies, focused on the morphology and ultrastructure of a ranavirus, showed that extracellular viral particles consist of a three-layered membrane including an external lipoprotein envelope, a protein capsid, and a lipid-containing membrane [13]. The authors suggested that the viral envelope derived from host plasma membranes by a budding process, and that it reacted with proteinase K, indicative of the protein content of the viral envelope [13]. This envelope enables the virus to enter neighboring cells by endocytosis to start a round infection.
Lysates of frog virus 3 (FV3) usually contain a mixture of naked nucleocapsids and enveloped particles. In fact, this virus can be released either by budding or by host cell lysis [4,6,18]. Braunwald et al. [3] reported that enveloped virions are more infective than naked ones, suggesting that the envelope could be a major factor in the early steps of virus-host interaction. By contrast, Heppell and Berthiaume [9] reported that FV3, chilo iridescent virus (CIV) and LCDV have a double-layered capsid, with a high proportion of phospholipids in the internal layer, and that an external envelope has not been observed. The authors suggested that damage during sample preparation might have accounted for the lack of a viral envelope in the virions.
González de Canales et al. [8] reported that, in LCDV-infected cells, at the end of the viral cycle, it is possible to observe a hyaline capsid partially composed of sulfate sialoglycoprotein surrounding the cellular membrane. Therefore, viral particles might acquire glycoproteins from this hyaline capsid during the budding process.
The presence of glycoproteins in LCDV particles has been confirmed by other authors. Robin et al. [15] detected at least ten glycoproteins in LCDV virions, one of which was identified as a surface glycoprotein; four that were located either on the surface or inside the virion; and the remaining glycoproteins found in a more internal position. These results differed from the general common observation that all viral glycoproteins are found at the surface of viral particles [14]. However, viral glycoproteins have also been detected inside the virion [5]. This glycoprotein location could be considered as viral components of intermediary steps of the viral cycle [1]. Based on the present study, it is not possible to confirm the internal or external location of these glycoproteins; however, the presence of a viral envelope suggests an external location and also that they might play a major role in virus-cell surface interactions.
Acknowledgements. This study was supported by a grant of CICYT of the Spanish Government (MAR99-0609). We greatly appreciate the assistance of Ms. M. J. Navarrete in the English version.
References
1. Basak S, Compans RW (1983) Studies on the role of glycosylation in the functions and antigenic properties of influenza virus glycoproteins. Virology128:77-91 [ Links ]
2. Bejar J, Borrego JJ, Alvarez MC (1997) A continuous cell line from the cultured marine fish gilt-head seabream (Sparus aurata L.). Aquaculture 150:143-153 [ Links ]
3. Braunwald J, Tripier F, Kirn A (1979) Comparison of the properties of enveloped and naked frog virus 3 (FV3) particles. J Gen Virol 45:673-682 [ Links ]
4. Braunwald J, Nonnenmacher H, Tripier-Darcy F (1985) Ultrastructural and biochemical study of Frog Virus 3 uptake by BHK-21 cells. J Gen Virol 66:283-293 [ Links ]
5. Caughman GB, Staczek J, O´ Callagham DJ (1984) Equine cytomegalovirus: structural proteins of virions and nucleocapsids. Virology 134:184-195 [ Links ]
6. Cuillel M, Tripier F, Braunwald J, Jacrot B (1979) A low resolution structure of Frog Virus 3. Virology 99:277-285 [ Links ]
7. Flügel RM, Darai G, Gelderblom H (1982) Viral proteins and adenosine triphosphate phosphorhydrolase activity of fish lymphocystis disease virus. Virology 122:48-55 [ Links ]
8. Gonzalez de Canales ML, Muñoz-Cueto JA, Arellano J, Garcia-Garcia A, Sarasquete C (1996) Histological and histochemical characteristics of the lymphocystis disease in gilthead seabream, Sparus aurata L. from the South Atlantic coast of Spain. Eur J Histochem 40:143-152 [ Links ]
9. Heppell J, Berthiaume L (1992) Ultrastructure of lymphocystis disease virus (LDV) as compared to frog virus 3 (FV3) and chilo iridiscent virus (CIV): effects of enzymatic digestions and detergent degradations. Arch Virol 125:215-226 [ Links ]
10. Kelly DC, Ayres MD, Lescott T, Robertson JS, Happ GM (1979) A small iridescent virus (type 29) isolated from Tenebrio molitor. A comparison of its proteins and antigens with six other iridescent viruses. J Gen Virol 42:95-105 [ Links ]
11. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685 [ Links ]
12. Paperna I, Sabnai I, Colorni A (1982) An outbreak of lymphocystis in Sparus aurata L. in the Gulf of Aqaba, Red Sea. J Fish Dis 5:433-437 [ Links ]
13. Qin QW, Lam TJ, Sin YM, Shen H, Chang SF, Ngoh GH, Chen CL (2001) Electron microscopic observations of a marine fish iridovirus isolated from brown-spotted grouper, Epinephelus tauvina. J Virol Meth 98:17-24 [ Links ]
14. Rifkin DB, Compans RW (1976) Identification of the spike proteins of Rous Sarcoma virus. Virology 46:485-489 [ Links ]
15. Robin J, Laperrière A, Berthiaume L (1986) Identification of the glycoproteins of lymphocystis disease virus (LDV) of fish. Arch Virol 87:297-305 [ Links ]
16. Sarasquete C, Gonzalez de Canales ML, Arellano J, Perez-Prieto S, Garcia-Rosado E, Borrego JJ (1998) Histochemical study of lymphocystis disease in skin of gilthead seabream, Sparus aurata L. Histol Histopathol 13:37-45 [ Links ]
17. Van Regenmortel MHV, Fauquet CM, Bishop DHL, Calisher CH, Carsten EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB (2000) Virus taxonomy: Classification and nomenclature of viruses. 7th Report of the ICTV. Academic Press, New York [ Links ]
18. Willis D, Granoff A (1974) Lipid composition of Frog Virus 3. Virology 61:256-269 [ Links ]