My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
International Microbiology
Print version ISSN 1139-6709
INT. MICROBIOL. vol.7 n.3 Sep. 2004
| RESEARCH ARTICLE | |||
|
| |||
| Functional characteristics of culturable bacterioplankton from marine and estuarine environments
Summary. Information on the structure of bacterioplankton communities is continuously increasing, while knowledge of their metabolic capabilities remains limited. In this study, the metabolic capacity of bacterioplankton was investigated, as such information is necessary to fully understand carbon cycling and other biogeochemical processes. The diversity of dominant culturable chemoorganotrophic bacteria from one estuarine and three marine environments was analyzed by random isolation of colony-forming units on solid media, taxonomical identification by partial 16S rRNA gene sequence analysis, and functional characterization of the isolates. A total of 76 16S rRNA gene sequences, representing 19 different genotypes, were obtained from the four sampling localities, including Bacillus, Pseudomonas, Pseudoalteromonas, Vibrio, and Erythrobacter as the most frequently isolated genera. The range of metabolic functions possessed by the cultured bacterial assemblages differed significantly between sites. Similarly, the percentage at each sampling station of bacteria capable of performing a specific function was significantly different for 18 of the 25 investigated metabolic functions. At two localities, the bacterial assemblages were dominated by a single genus (Pseudoalteromonas or Erythrobacter) and appeared to be functionally specialized. More than 95% of the isolates were capable of utilizing dissolved free amino acids and protein as their sole nitrogen sources, and all isolates of the specialized assemblages expressed β-glucosidase. Furthermore, only some of the isolates were able to utilize NH4+, while up to two thirds of the isolates of the two marine sites were able to grow on NO3-. [Int Microbiol 2004; 7(3):219-227] Key words: culturable bacterioplanktom · physiological characterization · taxonomical and functional diversity · metabolic capacity | ||
| |||
| Características funcionales del bacterioplancton cultivable de ambientes marinos y de estuario Resumen. La información sobre la estructura del bacterioplancton aumenta continuamente, mientras que el conocimiento de sus capacidades metabólicas sigue siendo limitado. En este estudio se investigó la capacidad metabólica del bacterioplancton, dado que dicha información es necesaria para comprender completamente el ciclo del carbono y otros procesos biogeoquímicos. La diversidad de las bacterias quimioorganotrofas cultivables que predominaban en un ambiente de estuario y en tres ambientes marinos se estudió aislando al azar unidades formadoras de colonias en medios sólidos, realizando la identificación taxonómica por medio del análisis de secuencias de genes del 16S rRNA, y mediante la caracterización funcional de los aislados. A partir de las cuatro localidades de muestreo se obtuvieron 76 secuencias de genes del 16S rRNA, que representaban 19 genotipos diferentes. Los géneros aislados con mayor frecuencia fueron Bacillus, Pseudomonas, Pseudoalteromonas, Vibrio y Erythrobacter. El margen de las funciones metabólicas que tenían los conjuntos (assemblages) de bacterias cultivadas difería notablemente entre las distintas localidades de muestreo. De manera similar, en cada estación de muestreo el porcentaje de bacterias que podían realizar alguna función específica era muy diferente para 18 de las 25 funciones metabólicas investigadas. En dos localidades predominaba un sólo género (Pseudoalteromonas o Erythrobacter) y parecían desempeñar funciones especializadas. Más del 95% de los aislados podían utilizar como única fuente de nitrógeno aminoácidos libres y proteínas disueltos, y todos los aislados de los conjuntos especializados expresaban β-glucosidasa. Además, sólo algunos de los aislados podían usar NH4+, mientras que hasta un tercio de los aislados en las dos localidades marinas podían crecer con NO3-. [Int Microbiol 2004; 7(3):219-227] Palabras clave: bacterioplancton cultivable · caracterización fisiológica · diversidad funcional y taxonómica · capacidad metabólica | Características funcionais do bacterioplâncton cultivável de ambientes marinhos e estuarinos Resumo. A informação sobre a estrutura do bacterioplâncton aumenta continuadamente enquanto que o conhecimento de sua capacidade metabólica continua limitado. Neste estudo se investigou a capacidade metabólica do bacterioplâncton, uma vez que tal informação é necessária para que se comprenda completamente o ciclo do carbono e outros processos biogeoquímicos. Foi estudada a diversidade das bactérias quimiorganotróficas cultiváveis que predominavam em um ambiente estuarino e em três marinhos isolando-se, aleatoriamente, unidades formadoras de colonias em meios sólidos e identificando-as taxonomicamente através da análise da seqüência de genes 16S rRNA e mediante a caracterização funcional dos isolados. A partir dos quatro locais amostrados foram obtidas 76 seqüências de genes 16S rRNA que representavam 19 genotipos diferentes. Os gêneros que foram isolados com maior freqüência foram: Bacillus, Pseudomonas, Pseudoalteromonas, Vibrio e Erythrobacter. A variação das funções metabólicas que tinha os conjuntos (assemblages) de bactérias cultivadas diferia notavelmente entre as distintas localidades amostradas. De maneira semelhante, em cada estação de amostragem, o percentual de bactérias que podia realizar alguma função específica era muito diferente para 18, das 25 funções metabólicas investigadas. Em dois locais, predominava um só gênero (Pseudoalteromonas ou Erythrobacter) os quais pareciam desempenhar funções especializadas. Mais de 95% dos isolados podiam utilizar aminoácidos e proteínas livres como única fonte de nitrogênio, e todos os isolados dos conjuntos especializados expressavam β-glucosidase. Somente alguns dos isolados podiam usar NH4+, enquanto que uma terceira parte, de duas localidades marinhas, podiam crescer com NO3-. [Int Microbiol 2004; 7(3):219-227] Palavras chave: bacterioplâncton cultivável · caracterização fisiológica · diversidade funcional e taxonômica · capacidade metabólica |
Introduction
Bacterioplankton plays a key role in the turnover of carbon and nitrogen in aquatic environments. Recent studies on the genetic diversity of bacterioplankton communities have shown taxonomic variation in both time and space [3,14]. However, since this information is based primarily on 16S rRNA gene sequences, the physiological characteristics and the ecological roles of the identified genotypes are largely unknown. Most of the currently available knowledge on bacterioplankton community function derives from "black box" studies in which turnover of dissolved organic matter by uncharacterized bacterial communities is measured [33], i.e. the role of the individual community member is unknown. A few studies, however, have tried to attribute specific functions to specific phylogenetic groups. For instance, Cottrell and Kirchman [10,11] showed, by combined microautoradiography and fluorescence in situ hybridization (FISH), that taxonomic groups at the division and subclass levels differed in their potential for utilizing organic compounds, and Davey et al. [13] demonstrated that differences in enzyme activity between communities could be explained by differences in species composition. Similarly, Cho and Giovannoni [8], in a study of the growth characteristics of marine oligotrophic gamma-proteobacteria under different culture conditions (temperature and carbon concentration), found that the physiological properties generally followed the phylogenetic relationships.
The aim of the present study was to assist in filling the knowledge gap between identity and function, i.e. to address the fundamental question of which functions are attributable to which species (genotype) in the natural environment. Hence, we carried out a comparative investigation of the relationship between the taxonomic and functional diversity (metabolic capacity) of bacterioplankton at the species level. The focus of the study was the culturable fraction of the bacterioplankton since a thorough physiological characterization requires bacteria in axenic culture [2,50]. Culture-dependent techniques (both traditional [6,26,32,40] and new high-throughput culturing methods [8,46]) have experienced a revival recently, and new evidence suggests that culture-independent and culture-dependent methods identify unique subsets of bacterial species [17,32,40]. Bacterial species potentially identified by culture-independent methods were, therefore, most likely overlooked by our approach. However, because the culturable fraction appears to represent the active populations [18,41,47], the isolated bacteria probably were of ecological significance at the time of sampling.
Materials and methods
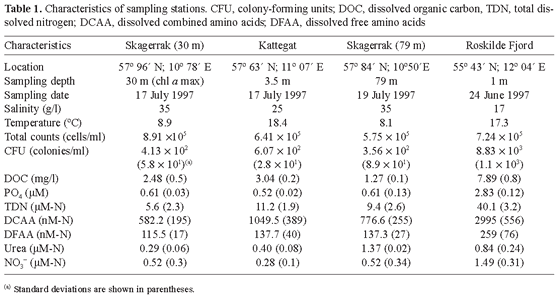
Sampling. Samples of Danish marine and estuarine water were collected at a depth of 3.5 m in the Kattegat, at 30 m (chlorophyll maximum) and 79 m in the Skagerrak, and at 1 m in the shallow estuary of Roskilde Fjord. Coordinates and characteristics of the sampling stations are listed in Table 1.
Chemical analyses. Water samples for analysis of o-phosphate, total dissolved nitrogen (TDN), nitrate, urea, dissolved free amino acids (DFAA), and dissolved combined amino acids (DCAA) were filtered through 0.2-µm cellulose acetate filters (Sartorius) and stored in acid-rinsed bottles at -18ºC. TDN was measured on a Dohrman TN1900 nitrogen analyzer using urea as standard. Nitrate, o-phosphate, and urea were determined on an AlpKem Flowsolution IV autoanalyzer according to procedures of the manufacturer, except urea, for which the method by Price and Harrison [44] was adopted. Concentrations of DFAA and DCAA (water samples hydrolyzed according to Jørgensen and Jensen [29]) were determined as fluorescent o-phthaldialdehyde derivates by HPLC according to the method of Jørgensen et al. [27].
For analysis of dissolved organic carbon (DOC), 0.2-µm cellulose-acetate-filtered samples were stored in closed precombusted glass sample vials (Shimadzu) at -18ºC. DOC was measured on a Shimadzu TOC 5000 analyzer (oxidation at 680ºC, Pt-covered aluminum oxide as a catalyst). At least triplicate injections were made per sample. Acidified solutions (Suprapur HCl, Merck, Darmstadt, Germany) of potassium biphthalate in Milli-Q water were used as standards.
Counting and isolation of bacteria. Samples for bacterial total counts were preserved with formaldehyde 2% (v/v) final concentration for Roskilde Fjord and 3.7% (v/v) final concentration for Skagerrak and Kattegat). Bacteria were counted under epifluorescence microscopy using acridine orange staining (Roskilde) or 4;6-diamidino-2-phenylindole (DAPI) staining (Skagerrak and Kattegat) according to the methods of Hobbie et al. [24] and Porter and Feig [43], respectively.
The number of colony-forming units (CFU) was determined by plate-spreading dilutions of filtered water (0.8-µm pore-size, Nuclepore polycarbonate filter) on 10% nutrient strength ZoBell agar plates [57]. The medium was diluted in order to reduce substrate-accelerated death. Plates contained 0.5 g Bacto peptone (Difco, Detroit, MI), 0.1 g yeast extract (Difco), 0.015 g FePO4, 4H2O (Riedel-de Haën AG, Seelze-Hannover, Germany) and 15 g agar (Difco) per liter of water from the appropriate sampling locality. The water had been sequentially filtered through 1.0- and 0.2-µm-pore-size filter cartridges (Millipore Corp., Bedford, MA), and a 0.2-µm GV filter (Nuclepore Corp., Pleasanton, CA). To insure that slow-growing strains were not overlooked, plates were incubated for 11 days at 16ºC (marine stations) or at room temperature (Roskilde Fjord). The incubation time of 11 days was based on a preliminary experiment (not shown) demonstrating that numbers of colonies appearing on the agar plates reached a constant value after 11-12 days.
Dilutions resulting in plates with ca. 30-50 colonies were preferentially used to isolate 30 colonies per sampling station. Randomness in colony selection was assured by observing agar plates in an orientation determined before the appearance of colonies, and by choosing colonies in a consistent manner, using horizontal gridlines. Rather than choosing different colonies, to obtain an estimate of species richness, the isolation procedure employed selected for abundant culturable bacteria and provided an estimate of their relative abundances. Most colonies were white, round and smooth, and a total of 16 different colony morphologies were observed. Isolates were stored frozen at -80ºC in 50% (v/v) glycerol in liquid 10% nutrient strength ZoBell medium.
Characterization of isolated bacteria. Isolated bacteria were grouped on the basis of their morphological and physiological characteristics. Isolates having the same colony morphology were often different with respect to their physiological characteristics. Gram reaction was determined by lysis with 3% (w/v) KOH [21]. Catalase activity was detected by the presence of bubbles after addition of one drop of 3% (v/v) H2O2 to colonies. Cytochrome oxidase activity tests were considered positive when cells formed blue pigments after being streaked on filter paper wetted with 1% (w/v) of N:N:N:N-tetramethyl-p-phenyl-diaminedihydrochloride. NO3--reduction, indole production from tryptophan, fermentation of glucose, L-arginine dihydrolase activity, urease activity, hydrolysis of gelatin, and growth on 12 different organic carbon compounds were determined using the API 20NE test system (bioMérieux, Marcy-l'Etoile, France) as recommended by the manufacturer.
The potential to grow on specific N sources was also assessed. Cells were grown for 2 days in liquid 10% strength ZoBell medium of appropriate salinity and starved for 4 days at 20ºC in sea salt (Sigma) or minimal medium [23] without an added carbon and nitrogen source. The starvation period was introduced in order to avoid positive signals (subsequent growth) resulting from utilization of internal carbon and nitrogen reserves. Following starvation, growth was tested in microtiter wells (Nunc, Naperville, IL) with 1 mM nitrogen derived from one of the following compounds as the sole nitrogen source: NH4Cl (Merck), NaNO3- (Merck), urea (Sigma, St. Louis, MO), a mixture of 22 amino acids (alanine, arginine hydrochloride, aspargine, aspartate, cysteine, cystine, glutamate, glutamine, glycine, histidine hydrochloride, trans-4-hydroxyproline, isoleucine, leucine, lysine hydrochloride, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, valine) (Sigma), casein (Sigma) or Escherichia coli DNA (Sigma). The nitrogen compounds were dissolved either in sea salt amended with 1 mM phosphate derived from KH2PO4 (Merck) and 100 mM carbon from D-glucose, H2O (Merck), or in minimal medium with glucose. The applied nutrient concentrations were chosen so that growth could be measured as an increase in optical density (OD590) on an ELISA reader EL314 (Bio-Tek instruments, Winooski, VT). Wells with nitrogen but no cells, and wells with cells but no nitrogen served as controls. Growth was followed for 5 days. Glassware and pipette tips were acid rinsed to avoid contamination with nitrogen. An isolate was considered capable of growth on a specific nitrogen source if growth occurred in at least two out of three or three out of four replicate tests. From each locality, 28 to 29 of the isolates were characterized with respect to enzymatic activities and carbon assimilation (API 20NE test system), while 18 to 22 isolates were characterized with respect to nitrogen assimilation in the microtiter wells. The remaining isolates failed to resume growth after storage.
16S rDNA sequencing. DNA was extracted by transferring colonies with a sterile toothpick into 1.5-ml safe-lock Eppendorf tubes containing 200 µl 0.04 M Tris-acetate EDTA buffer (Sigma). Tubes were vortexed, boiled for 10 min, left at room temperature for 10 min, and centrifuged (15,000 g, 10 min, 4ºC). Supernatants containing the DNA were then transferred to new tubes and stored at 5ºC.
DNA coding for 16S rRNA was amplified by polymerase chain reaction (PCR) (10 min at 95ºC; 35 cycles of 30 s at 95ºC, 30 s at 55ºC, 2 min at 72ºC; 6 min at 72ºC; final cooling at 4ºC). The PCR protocol of Johnsen et al. [25] was applied, except that (i) Ampli Taq Gold polymerase (Applied Biosystems, Foster City, CA.), and (ii) primers F1 at position 9-27 and R13 at position 1525-1542 [15] were used. After PCR, products were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. A partial sequence of the 16S rDNA was obtained by sequencing (ABI PRISM 3700 DNA Analyzer, BigDye Terminator, Applied Biosystems) the PCR product in both directions using primer F1 and a primer (CGTATTACCGCGGCTGCT) modified after the method of Lane et al. [34] and binding at position 518-535. Sequences were aligned against those from the Ribosomal Database Project II [35].
Approximately two-thirds of the 30 strains from each sampling station were sequenced. The remaining isolates either were unable to grow after long-term storage or failed in producing a PCR product or a 16S rDNA sequence.
Accession numbers. Nucleotide sequences were filed in GenBank under the following accession numbers: A5: AF293089; A6: AF293090; A7: AF325137; A10: AF325138; A22: AF325139; A25: AF325135; A28: AF293091; B3: AF293092; B4: AF293093; B20: AF325140; B25: AF325141; B30: AF325142; D1: AF293094; D3: AF325143; D6: AF325144; D15: AF325146; D23: AF325147; D25: AF293095; D30: AF325148; F15: AF325136; F16: AF325149; F19: AF325150. Identical clone sequences were not filed.
Statistical analysis. The ability of a bacterial isolate to perform a specific metabolic function (e.g. ability to grow on casein) was treated as a binary response, i.e. the population of bacterial isolates being able to perform a metabolic function was binomially distributed with probability parameter "p" and sample size "n". The data were analyzed using logistic regression under the framework of generalized linear models [37]. A likelihood-ratio test was used to determine whether the frequency of an individual metabolic function, or the frequencies of all metabolic functions combined, were significantly different between sampling stations. Models applied were Npositive/Ntotal = station and Npositive/Ntotal = stationi + functional groupj + stationi× functional groupj + functionn (functional groupj), respectively. The logistic regressions were carried out using PROC GENMOD(SAS/STAT). A χ2 test was used to test whether the total numbers of functions present at each station were significantly different. A phenogram (dendrogram) based on the characterization with respect to enzymatic capabilities and growth on various carbon and nitrogen sources was constructed using the Treecon software [53]. A bootstrap analysis with 500 sample replications was performed using the simple matching similarity index and the UPGMA algorithm.
Results
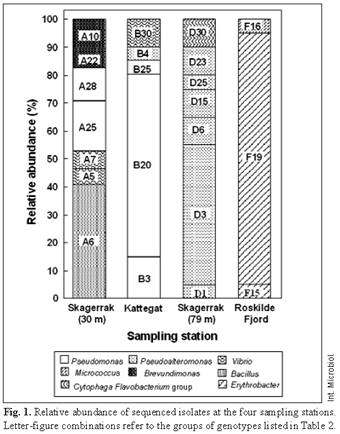
Diversity of isolated bacteria. A total of 76 partial 16S rRNA gene sequences (genotypes) were obtained from the four sampling sites. Of these, 19 were found to be different (Table 2). In three cases, identical sequences were obtained at different sampling stations: Isolate B4 was identical to D3, A28 was identical to B20, and A25 was identical to B3 (Table 2). Isolates with the B4/D3 sequence were most abundant at Skagerrak (79 m) but were also found in Kattegat (Fig. 1, Table 2). These isolates had a 100% partial 16S rDNA similarity with Pseudoalteromonas haloplanktis SKA19 previously isolated from the Skagerrak (Table 2). The two other pairs of isolates (A25/B3 and A28/B20) were found in the Skagerrak (30 m) and Kattegat (Fig. 1, Table 2) and were closely related to Pseudomonas sp. OA146, which is a salt-adapted pseudomonad isolated from the Baltic Sea that synthesizes glucosylglycerol as an osmolyte [38].
The number of different genotypes within each locality varied between three and seven (Fig. 1). At both Skagerrak sites, seven different 16S rRNA gene sequences were identified (Fig. 1). Bacillus spp. and pseudomonads dominated in the Skagerrak (30 m), constituting 41% and 29% of the sequenced isolates, respectively (Fig. 1, Table 2), while pseudoalteromonads made up 90% of the sequenced isolates at Skagerrak (79 m) (Fig. 1, Table 2). In the Kattegat, five different 16S rRNA gene sequences were obtained (Fig. 1), with Pseudomonas being the dominant genus (85%) (Fig. 1, Table 2). At the estuarine station in Roskilde Fjord, 18 out of 19 isolates were identified as Erythrobacter and only three different genotypes were found (Fig. 1, Table 2).
The 16S rRNA gene sequence similarity between 17 of the different isolates and previously reported sequences obtained from geographical distant localities was higher than 99% (Table 2). The two isolates (D23 and D30) that were less than 99% similar to literature sequences were both found in the Skagerrak (79 m). Isolate D30 has recently been described as a new species within the genus Tenacibaculum [19].
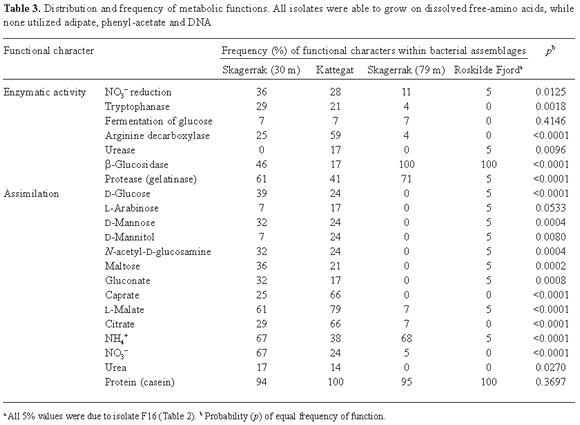
Functional characteristics of isolated bacteria. The number of metabolic functions possessed by the isolated assemblages of bacteria from the four sampling localities was significantly different (p = 0.0025). Similarly, the percentage at each sampling station of bacteria capable of performing a specific function differed significantly (p < 0.05) for 18 of the 25 investigated metabolic functions (Table 3).
The Kattegat and Skagerrak (30 m) isolates appeared to be more versatile than those of Roskilde Fjord and Skagerrak (79 m) as 88% and 85% of the investigated metabolic characteristics were expressed at these localities, respectively, while only 62% and 50% were found in Roskilde Fjord and Skagerrak (79 m), respectively. Furthermore, specific metabolic functions were expressed either by all or by none of the members of the Roskilde Fjord and Skagerrak (79 m) assemblages, whereas in Skagerrak (30 m) and Kattegat the ability to perform a metabolic function was more evenly shared among the isolates (Table 3). A cluster analysis (Fig. 2) based on the functional characteristics of the isolates demonstrated that the Roskilde Fjord and Skagerrak (79 m) assemblages clustered in relatively distinct groups (except for isolate F16), whereas no distinct functional clusters were observed for Kattegat and Skagerrak (30 m).
More than 95% of the isolates were capable of utilizing DFAA and protein as sole nitrogen sources, and at Skagerrak (79 m) and Roskilde Fjord all isolates expressed β-glucosidase activity (Table 3). Surprisingly, not all isolates were able to utilize NH4+ as sole N source, and two thirds of the isolates in Skagerrak (30 m) and about 40% of those in Kattegat were able to grow on NO3-. None of the isolates grew on DNA, adipate or phenyl acetate (Table 3).
Discussion
The taxonomy and potential metabolic capacity of culturable bacterioplankton from one estuarine and three marine localities were studied. The metabolic potential rather than the in situ activity of the isolates was determined since in situ measurements are only representative of the conditions at the time of sampling. Assessment of the potential metabolic capacity, therefore, provides a better picture of the ecological roles of the organisms. The bacterial isolates from the Skagerrak (79 m) were dominated by a single genus (Pseudoalteromonas) and were functionally less versatile than those in Kattegat and Skagerrak (30 m) (Fig. 1, Table 2). Consistent with this, all isolates tested positive for β-glucosidase activity, while only a few were able to grow on low-molecular-weight carbon substrates (Table 3). If the culturable bacteria represent those that are active in the environment, as suggested by Ellis et al. [18], Pinhassi et al. [41] and Rehnstam et al. [47], this suggests that the Skagerrak (79 m) community was adapted to growth on refractory carbon possibly in the form of aged organic matter. Roskilde Fjord isolates were almost completely dominated by a genotype closely related to Erythrobacter longus OCh101, which has been frequently isolated from eutrophic environments [49]. Similar to the isolates in Skagerrak (79 m), only a few of those from Roskilde Fjord were able to grow on low molecular weight substrates while they all showed β-glucosidase activity (Table 3), suggesting that the community of culturable bacteria was adapted to growth on refractory (allochthonous) carbon. Bacillus and Pseudomonas spp. dominated the Skagerrak (30 m) and Kattegat samples (Fig. 1, Table 2). The Bacillus isolates were capable of metabolizing a diverse range of carbohydrates. Similarly, pseudomonads have simple nutritional requirements and grow well in media containing organic matter in solution [39]. Thus, the bacteria at the chlorophyll a maximum in the Skagerrak (30 m) and in the surface water in Kattegat probably grew on labile dissolved organic matter produced by the phytoplankton [9].
All, or nearly all, of the isolated bacteria were able to utilize dissolved free and combined amino acids as their sole nitrogen sources (Table 3). In contrast, not all isolates were able to utilize NH4+. This was surprising since NH4+ is generally considered to be an important source of N for bacterioplankton [33]. Equally surprising was the fact that two thirds of the isolates in Skagerrak (30 m) and about 40% of those in Kattegat were able to grow on NO3- (Table 3) since nitrate is not considered to be a major contributor to bacterial nitrogen requirements [56]. No isolates grew on DNA as the sole nitrogen source (Table 3) suggesting that it served primarily as a supplementary source of nitrogen [28] or phosphorus [52]. Also, studies have shown that bacterioplankton involved in cycling of dissolved DNA preferably take up small-size DNA fragments (100 bp) [27]. In the present study, the size of the DNA fragments might have been too large. Only a few of the bacterial isolates could grow on urea (Table 3). This is consistent with results in the literature; for example, Cho et al. [7] reported that bacterioplankton are significant urea producers but not decomposers, and Jørgensen et al. [30] found that urea was produced when amino acids were decomposed.
In order to advance our understanding of carbon cycling and other biogeochemical processes in aquatic ecosystems, detailed studies on the functional characteristics of bacterioplankton are needed. The results reported in this study expand the knowledge of the functional roles of marine pelagic bacteria. Currently, the best way to obtain in-depth information on growth characteristics and enzymatic capabilities at the species level is to study cultured bacteria. However, to overcome the potential biases of cultivation-dependent approaches, and to link measurements of potential metabolic capability with those of in situ activity, new methods, such as DNA/RNA-based stable-isotope probing [45], should be developed and applied in conjunction with classical ones.
Acknowledgements. We thank H. Irming, L. Hemmingsen, R. Elhøj Jensen and A. S. B. Hentze for technical assistance, the crew on DANA for help with water collection, J. Carstensen for performing the statistical analyses, and P. H. Pritchard and A. Johnsen for reviewing the manuscript. The work was supported by the Danish Natural Sciences Research Council (grants 9601731 and 9701015), and Center for Biological Processes in Contaminated Soil and Sediment.
References
1. Abraham W-R, Strömpl C, Meyer H, Lindholst S, Moore ERB, Christ R, Vancannret M, Tindall BJ, Bennasar A, Smit J, Tesar M (1999) Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int J Syst Bacteriol 49:1053-1073 [ Links ]
2. Achenbach LA, Coates JD (2000) Disparity between bacterial phylogeny and physiology. ASM News 66:714-715 [ Links ]
3. Acinas SG, Rodríguez-Valera F, Pedrós-Alió C (1997) Spatial and temporal variation in marine bacterioplankton diversity as shown by RFLP fingerprinting of PCR-amplified 16S rDNA. FEMS Microbiol Ecol 24:27-40 [ Links ]
4. Arrieta JM, Weinbauer MG, Herndl GJ (2000) Interspecific variability in sensitivity to UV radiation and subsequent recovery in selected isolates of marine bacteria. Appl Environ Microbiol 66:1468-1473 [ Links ]
5. Boivin-Jahns V, Bianchi A, Ruimy R, Garcin J, Daumas S, Christen R (1995) Comparison of phenotypical and molecular methods for the identification of bacterial strains isolated from a deep subsurface environment. Appl Environ Microbiol 61:3400-3406 [ Links ]
6. Brinkmeyer R, Knittel K, Jürgens J, Weyland H, Amann R, Helmke E (2003) Diversity and structure of bacterial communities in Arctic versus Antarctic pack ice. Appl Environ Microbiol 69:6610-6619 [ Links ]
7. Cho J-C, Park MG, Shim JH, Azam F (1996) Significance of bacteria in urea dynamics in coastal surface waters. Mar Ecol Prog Ser 142:19-26 [ Links ]
8. Cho JC, Giovannoni SJ (2004) Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol 70:432-440 [ Links ]
9. Cole JJ, Findlay S, Pace ML (1988) Bacterial production in fresh and saltwater ecosystems:a cross-system overview. Mar Ecol Prog Ser 43:1-10 [ Links ]
10. Cottrell MT, Kirchman DL (2000a) Natural assemblages of marine Proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl Environ Microbiol 66:1692-1697 [ Links ]
11. Cottrell MT, Kirchman DL (2000b) Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol 66:5116-5122 [ Links ]
12. Crump BC, Armbrust EV, Baross JA (1999) Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl Environ Microbiol 65:3192-3204 [ Links ]
13. Davey KE, Kirby RR, Turley CM, Weightman AJ, Fry JC (2001) Depth variation of bacterial extracellular enzyme activity and population diversity in the northeastern North Atlantic Ocean. Deep-Sea Res Part II: Topical Studies in Oceanography 48:1003-1017 [ Links ]
14. Dojka MA, Harris JK, Pace NR (2000) Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl Environ Microbiol 66:1617-1621 [ Links ]
15. Dorsch M, Stackebrandt E (1992) Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J Microbiol Methods 16:271-279 [ Links ]
16. Drancourt M, Bollet C, Carlioz A, Martelin R, Gayral J-P, Raoult D (2000) 16S Ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 38:3623-3630 [ Links ]
17. Eilers H, Pernthaler J, Glöckner FO, Amann R (2000) Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl Environ Microbiol 66:3044-3051 [ Links ]
18. Ellis RJ, Morgan P, Weightman AJ, Fry JC (2003) Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol 69:3223-3230 [ Links ]
19. Frette L, Jørgensen NOG, Irming H, Kroer N (2004) Tenacibaculum skagerrakense sp. nov., a marine bacterium isolated from the pelagic zone in Skagerrak, Denmark. Int J Syst Evol Microbiol 54:519-524 [ Links ]
20. Gauthier G, Gauthier M, Christen R (1995) Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int J Syst Bacteriol 45:755-761 [ Links ]
21. Gregersen T (1978) Rapid method for distinction of gram-negative from gram-positive bacteria. Eur J Appl Microbiol 5:123-127 [ Links ]
22. Hagström Å, Pinhassi J, Zweifel UL (2000) Biogeographical diversity among marine bacterioplankton. Aquat Microb Ecol 21:231-244 [ Links ]
23. Hareland WA, Crawford RL, Chapman PJ, Dagley S (1975) Metabolic function and properties of 4-hydroxyphenylacetic acid 1-hydrolase from Pseudomonas acidovorans. J Bacteriol 121:272-285 [ Links ]
24. Hobbie JE, Daley RJ, Jasper S (1977) Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol 33:1225-1228 [ Links ]
25. Johnsen K, Enger Ø, Jacobsen CS, Thirup L, Torsvik V (1999) Quantitative selective PCR of 16S ribosomal DNA correlates well with selective agar plating in describing population dynamics of indigenous Pseudomonas spp. in soil hot spots. Appl Environ Microbiol 65:1786-1788 [ Links ]
26. Junge K, Imhoff JF, Staley T, Deming JW (2002) Phylogenetic diversity of numerically important Arctic sea-ice bacteria cultured at subzero temperature. Microb Ecol 43:315-328 [ Links ]
27. Jørgensen NOG, Kroer N, Coffin RB, Yang X-H, Lee C (1993) Dissolved free amino acids, combined amino acids, and DNA as sources of carbon and nitrogen to marine bacteria. Mar Ecol Prog Ser 98:135-148 [ Links ]
28. Jørgensen NOG, Kroer N, Coffin RB (1994) Utilization of dissolved nitrogen by heterotrophic bacterioplankton: Effects of substrate C/N ratio. Appl Environ Microbiol 60:4124-4133 [ Links ]
29. Jørgensen NOG, Jensen RE (1997) Determination of dissolved combined amino acids using microwave-assisted hydrolysis and HPLC precolumn derivatization for labeling of primary and secondary amines. Mar Chem 57:287-297 [ Links ]
30. Jørgensen NOG, Kroer N, Coffin RB, Hoch MB (1999). Relations between bacterial nitrogen metabolism and growth efficiency in an estuarine and an open-water ecosystem. Aquat Microb Ecol 18:247-261 [ Links ]
31. Kaye JZ, Baross JA (2000) High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol Ecol 32:249-260 [ Links ]
32. Kisand V, Wikner J (2003) Combining culture-dependent and -independent methodologies for estimation of richness of estuarine bacterioplankton consuming riverine dissolved organic matter. Appl Environ Microbiol 69:3607-3616 [ Links ]
33. Kroer N, Jørgensen NOG, Coffin RB (1994) Utilization of dissolved nitrogen by heterotrophicbacterioplankton. A comparison of three ecosystem. Appl Environ Microbiol 60:4116-4123 [ Links ]
34. Lane DL, Pace B, Olsen GJ, Stahl DA, Sorgin ML, Pace NR (1985) Rapid determination of 16S rRNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA 82:6955-6955 [ Links ]
35. Maidak BL, Cole JR, Lilburn TG, Parker Jr CT, Saxman PR, Stredwick JM, Garrity GM, Li B, Olsen GJ, Pramanik S, Schmidt TM, Tiedje JM (2000) The RDP (Ribosomal Database Project) continues. Nucleic Acids Res 28:173-174 [ Links ]
36. Maruyama A, Honda D, Yamamoto H, Kitamura K, Higashihara T (2000) Phylogenetic analysis of psychrophilic bacteria isolated from the Japan Trench, including a description of the deep-sea species Psychrobacter pacificencis sp. nov. Int J Syst Bacteriol 50:835-846 [ Links ]
37. McCullagh P, Nelder JA (1989) Generalized linear models. 2nd ed. Chapman & Hall/CRC, Boca Raton, Florida, USA [ Links ]
38. Mikkat S, Galinski EA, Berg G, Minkwitz A, Schoor A (2000) Salt adaption in pseudomonads: characterization of glucosylglycerol-synthesizing isolates from brackish coastal waters and the rhizosphere. Syst Appl Microbiol 23:31-40 [ Links ]
39. Palleroni NJ (1991) Introduction to the familly Pseudomonadaceae. In: Balows et al. (eds.) The Prokaryotes, Vol. III, 2nd Ed, Springer-Verlag, New York, pp.: 3071-3085 [ Links ]
40. Pearce DA, van der Gast CJ, Lawley B, Ellis-Ewans JC (2003) Bacterioplankton community diversity in a maritime Antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS Microbiol Ecol 45:59-70 [ Links ]
41. Pinhassi J, Zweifel UL, Hagström Å (1997) Dominant marine bacterioplankton species found among colony-forming bacteria. Appl Environ Microbiol 63:3359-3366 [ Links ]
42. Pinhassi J, Azam F, Hemphälä J, Long RA, Martinez J, Zweifel UL, Hagström Å (1999) Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat Microb Ecol 17:13-26 [ Links ]
43. Porter KG, Feig YS (1980) The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr 25:943-948 [ Links ]
44. Price NM, Harrison PJ (1987) Comparison of methods for analysis of dissolved urea in seawater. Mar Biol 94:307-317 [ Links ]
45. Radajewski S, McDonald IR, Murrell JC (2003) Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr Opin Biotechnol 14:296-302 [ Links ]
46. Rappé MS, Connon SA, Vergin KL, Giovannoni SJ (2002) Cultivation of the ubiquitous SAR 11 marine bacterioplankton clade. Nature 418:630-633 [ Links ]
47. Rehnstam AS, Bäckman S, Smith DC, Azam F, Hagström Å (1993) Blooms of sequence-specific culturable bacteria in the sea. FEMS Microbiol Ecol 102:161-166 [ Links ]
48. Sawabe T, Tanaka R, Iqbal MM, Tajima K, Enzura Y, Ivanova EP, Christen R (2000) Assignment of Alteromonas elyakovii KMM 162T and five strains isolated from spot-wounded fronds of Laminaria japonica to Pseudoalteromonas elyakovii comb. nov. and the extended description of the species. Int J Syst Evol Microbiol 50:265-271 [ Links ]
49. Shiba T, Simidu U (1982) Erythrobacter longus gen. nov., sp. nov., an aerobic bacterium which contains bacteriochlorophyll a. Int J Syst Bacteriol 32:211-217 [ Links ]
50. Stackebrandt E, Tindall BJ (2000) Appreciating microbial diversity: rediscovering the importance of isolation and characterization of microorganisms. Environ Microb 2:9-10 [ Links ]
51. Suzuki M, Nakagawa Y, Harayama S, Yamamoto S (2001) Phylogenetic analysis and taxonomic study of marine Cytophaga-like bacteria: proposal for Tenacibaculum gen. nov. with Tenacibaculum maritimum comb. nov. and Tenacibaculum ovolyticum comb. nov., and description of Tenacibaculum mesophilum sp. nov. and Tenacibaculum amylolyticum sp. nov. Int J Syst Evol Microbiol 51:1639-1652 [ Links ]
52. Turk V, Rehnstam A-S, Lundberg E, Hagström Å (1992) Release of bacterial DNA by marinenanoflagellates, an intermediate step in phosphorus regeneration. Appl Environ Microbiol 58:3744-3750 [ Links ]
53. van de Peer Y, de Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569-570 [ Links ]
54. Verhille S, Baida N, Dabboussi F, Izard D, Leclerc H (1999) Taxonomic study of bacteria isolated from natural mineral waters: proposal of Pseudomonas jessenii sp. nov. and Pseudomonas mandelii sp. nov. Syst Appl Microbiol 22:45-58 [ Links ]
55. Vybiral D, Denner EBM, Haller CH, Busse H-J, Witte A, Höfle MG, Velimirov, B (1999) Polyphasic classification of 0.2 µm filterable bacteria from the Western Mediterranean Sea. Syst Appl Microbiol 22:635-646 [ Links ]
56. Wheeler PA, Kirchman DL (1986) Utilization of inorganic and organic nitrogen by bacteria in marine systems. Limnol Oceanogr 31:998-1009 [ Links ]
57. ZoBell CE (1946) Marine microbiology: a monograph on hydrobacteriology. Chronica Botanica Co, Waltham, MA, USA [ Links ]