Mi SciELO
Servicios Personalizados
Revista
Articulo
Indicadores
-
 Citado por SciELO
Citado por SciELO -
 Accesos
Accesos
Links relacionados
-
 Citado por Google
Citado por Google -
 Similares en
SciELO
Similares en
SciELO -
 Similares en Google
Similares en Google
Compartir
International Microbiology
versión impresa ISSN 1139-6709
INT. MICROBIOL. vol.9 no.1 mar. 2006
RESEARCH NOTE
Genetic diversity and recombination within populations of Fusarium pseudograminearum from western Canada
Prashant K. Mishra1; Jalpa P. Tewari1; Randall M. Clear2; T. Kelly Turkington3
1Department of Agricultural, Food, and Nutritional Science, University of Alberta, Edmonton, Alberta, Canadan
2Canadian Grain Commission, Winnipeg, Manitoba, Canada
3Lacombe Research Centre, Agriculture and Agri-Food Canada, Lacombe, Alberta, Canada
SUMMARY
Genetic diversity within populations of Fusarium pseudograminearum isolated from wheat grains from the Canadian provinces of Alberta and Saskatchewan was investigated. Three restriction enzymes (EcoRI, HaeIII, and PstI) were used to carry out restriction analysis of the nuclear ribosomal DNA (nrDNA) intergenic spacer region (IGS region) and eight primers were used to generate inter-simple sequence-repeat (ISSR) molecular markers. Our study indicated substantially high genetic diversity within these two populations, but low genetic differentiation and frequent gene flow among populations. The IGS data showed no genetic distinction between the two Alberta populations and only minor genetic differentiation between the Saskatchewan and Alberta populations. Analysis of molecular variance indicated that most genetic variability resulted from differences among isolates within populations. Multilocus linkage disequilibrium analysis suggested a panmictic population genetic structure and the occurrence of significant recombination in F. pseudograminearum. Regular gene flow and random mating between isolates from different populations could result in novel genotypes with both improved pathological and biological traits. [Int Microbiol 2006; 9(1):65-68].
Key words: Fusarium pseudograminearum · Gibberella coronicola · genetic diversity · gene flow · genetic recombination
RESUMEN
Diversidad genética y recombinación en poblaciones de Fusarium pseudograminearum del oeste de Canadá
Se investigó la diversidad genética en poblaciones de Fusarium pseudograminearum aisladas de semillas de trigo de las provincias canadienses de Alberta y Saskatchewan. Se usaron tres enzimas de restricción (EcoRI, HaeIII, and PstI) para analizar los marcadores moleculares de las regiones espaciadoras intergénicas (IGS) del DNA ribosómico del núcleo (nrDNA) y de los fragmentos generados entre repeticiones de secuencias sencillas (fragmentos ISSR). Nuestro estudió reveló una gran diversidad genética en ambas poblaciones, pero poca diferenciación genética y un flujo genético frecuente entre las poblaciones. Los datos relativos a las IGS no mostraron diferencias genéticas entre las dos poblaciones de Alberta estudiadas y sólo una ligera diferenciación entre las poblaciones de Alberta y de Saskatchewan. El análisis de la varianza molecular indicó que la variabilidad genética respondía en su mayor parte a diferencias entre aislamientos dentro de las poblaciones. El análisis del desequilibrio en el ligamiento genético sugería una estructura genética de la población de tipo panmíctico y la existencia de una recombinación significativa en F. pseudograminearum. Un flujo genético regular y el apareamiento al azar de aislamientos de poblaciones distintas podría producir nuevos genotipos con características biológicas mejores o patológicas. [Int Microbiol 2006; 9(1):65-68].
Palabras clave: Fusarium pseudograminearum · Gibberella coronicola · diversidad genética · flujo genético · recombinación genética
RESUMO
Diversidade genética e recombinação em populações de Fusarium pseudograminearum do oeste do Canadá
Foi averiguada a diversidade genética em populações de Fusarium pseudograminearum isoladas de sementes de trigo das províncias candenses de Alberta e Saskatchewan. Foram utilizadas três enzimas de restrição (EcoRI, HaeIII, and PstI) para analisar os marcadores moleculares das regiões espaçadoras intergênicas (IGS) do DNA ribossômico nuclear (nrDNA) e dos fragmentos gerados entre repetições de seqüências simples (fragmentos ISSR). Nosso estudou revelou uma grande diversidade genética em ambas populações, mas pouca diferenciação genética e um fluxo genético freqüente entre as populações. Os dados relativos às IGS no mostraram diferenças genéticas entre as duas populações de Alberta estudadas e apenas uma leve diferenciação entre as populações de Alberta e de Saskatchewan. A análise da variânza molecular indicou que a variabilidade genética correspondia em sua maioria a diferenças entre isolados de uma mesma população. A análise do desequilíbrio de ligação gênica sugeriu uma estrutura genética da população de tipo panmítico e a existência de uma recombinação significativa em F. pseudograminearum. Um fluxo gênico regular e o apareamiento ao acaso de isolamentos de populações diferentes poderia produzir novos genotipos com características biológicas melhores ou patológicas. [Int Microbiol 2006; 9(1):65-68].
Palavras chave: Fusarium pseudograminearum · Gibberella coronicola · diversidade genética · fluxo genético · recombinação genética
Introduction
Fusarium pseudograminearum Aoki and O'Donnell (teleomorph: Gibberella coronicola Aoki and O'Donnell), formerly known as F. graminearum group 1, is a haploid, heterothallic fungal pathogen belonging to Fusarium section Discolor [2]. Over the past few years, F. pseudograminearum has emerged as a major causal agent of crown-rot disease in cereal crops in various parts of the world [5]. The infection of cereal crops with F. pseudograminearum can result in a reduction in both grain yield and grain quality due to contamination with mycotoxins [4]. This species of Fusarium was identified in Canada only recently, and it has generally been isolated from the cereal-growing areas of western Canada. Isozyme analysis of F. pseudograminearum showed identical fingerprints among isolates [8], whereas phylogenetic analysis, using DNA sequence data from coding regions, demonstrated two genetic groups among the four isolates examined [15]. Although these studies provided useful preliminary information regarding the genetic structure of this fungus, there is still a need for further investigations to elucidate the genetic diversity and evolution of F. pseudograminearum.
The aim of the present study was to examine the genetic diversity within populations of F. pseudograminearum collected from infected wheat seed grown in the western Canadian provinces of Alberta and Saskatchewan. Restriction analysis of the nuclear ribosomal DNA (nrDNA) intergenic spacer region (IGS) and inter-simple sequence-repeat (ISSR) molecular markers were employed to analyze the genetic diversity and evolutionary dynamics of F. pseudograminearum. Both of these molecular markers have been used extensively to study the population genetics of various eukaryotes and have provided important genetic information relevant to the resolution of various genetic and evolutionary properties in a range of fungi [7,12,18,19].
Materials and methods
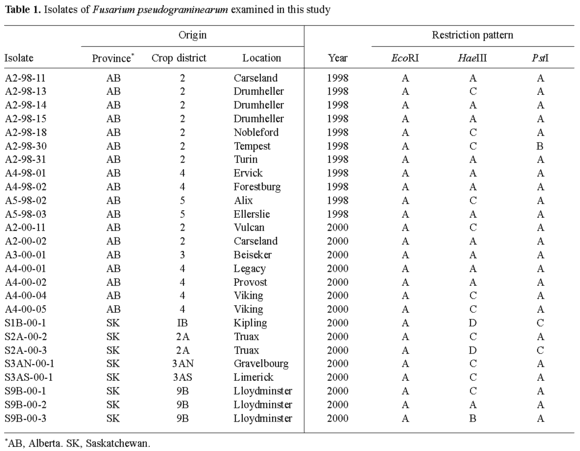
Fungal isolates and DNA extraction. Twenty-six isolates of F. pseudograminearum were isolated from infected wheat grains collected from the western Canadian provinces of Alberta and Saskatchewan (Table 1). The single-spored isolates were grown on potato-dextrose agar (PDA) plates at room temperature for 7 days. The tissue was harvested from the isolates grown on PDA, ground in liquid nitrogen, and DNA was extracted using the procedure described previously [9]. The identity of the isolates was established using a PCR-based approach that had been developed for F. pseudograminearum [2].
DNA fingerprinting and gel electrophoresis. The nrDNA IGS region was amplified from all isolates using primers, reagents, and conditions as described previously [11]. From each isolate, 10 µl of the amplified IGS product was digested with three different restriction enzymes (EcoRI, HaeIII, and PstI) in separate reactions carried out according to the manufacturer's recommendations (Invitrogen, Canada). The digests were size-fractionated through a 2% agarose gel containing ethidium bromide, visualized, and photographed under ultraviolet light using a Gel Documentation System (Kodak, Canada). The resulting IGS fingerprints were examined to identify the number of haplotypes among the tested isolates; based on these results, seven isolates representing different IGS haplotypes were selected for ISSR analysis (Table 1). ISSR fingerprinting was carried out using the procedures and the eight ISSR primers described in our previous study [13]. The ISSR analysis was repeated and bands that appeared in both amplifications were regarded as markers in this study. The resulting fingerprints from both (IGS and ISSR) analyses were scored as presence (1) or absence (0) of the bands for each isolate.
Analysis of the molecular data. Genetic diversity within a population was calculated from the binary data using the average gene diversity over loci [13]; the genotypic diversity, using the equation described previously [21]; and the level of polymorphism. Fixation index (FST), and gene flow (Nm) estimates [16,20] were used to infer genetic differentiation between populations. Analysis of molecular variance (AMOVA) was applied to partition the genetic diversity into, among, and within population components [6]. All statistical measures were computed using the software Arlequin, version 2.0, a software for population genetic data analysis [University of Geneva, Switzerland]. Recombination in F. pseudograminearum was estimated using the rd statistic as installed in the software MultiLocus [1]. The rd statistic is based on the variance of the genetic distance between pairs of isolates, compared to the corresponding variance in null data created through randomization of observed data under the expectation of random mating. In the present study, molecular marker data were randomized 1000 times to create a null distribution against which the observed rd value was compared. The observed rd statistic for a recombining organism is expected to be non-significant from the rd statistic inferred from the null data [1].
Results and Discussion
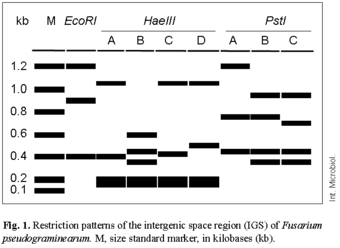
Restriction analysis of the IGS region resulted in 18 markers, of which 12 were polymorphic, whereas ISSR analysis resulted in 120 markers, of which 118 were polymorphic characters. Restriction analysis of the PCR-amplified IGS products resulted in five unique haplotypes among 26 isolates examined (Fig. 1, Table 1). The genotypic diversity calculated from the IGS data was relatively high in all populations, with the highest (0.980) found in the Alberta 2000 populations, followed by the Saskatchewan 2000 and Alberta 1998 populations, with genotypic diversities of 0.727 and 0.683, respectively. The genotypic diversity calculated from the ISSR data was 1.0, as all the isolates examined revealed different banding patterns. The average gene diversity over loci, inferred from the IGS data, was lower in population samples from Alberta (mean ± standard deviation: Alberta 1998, 0.079 ± 0.153; Alberta 2000, 0.054 ± 0.158) than in those from Saskatchewan (0.213 ± 0.173), whereas the ISSR results indicated similar values for samples from the two provinces (Alberta 1998, 0.394 ± 0.297; Saskatchewan 2000, 0.370 ± 0.245). However, there were no significant differences in the average gene diversity over loci among populations for both of the datasets, as was evident from the large standard deviations. The AMOVA results demonstrated that most of the genetic variation (94.17%) in the IGS region of F. pseudograminearum was due to within-population variability, while only 5.83% of the total variation occurred among populations. The within- and among-population variabilities for the ISSR data were 88.42% and 11.58%, respectively. The genetic diversity and the partitioning of genetic variation in F. pseudograminearum populations observed in our study are in agreement with the estimates of genetic diversity reported by other researchers for closely related Fusarium species, such as F. graminearum [10,13,21]. There are substantial similarities in the pathological, physiological, and ecological properties of the different species of Fusarium causing diseases in cereals [3]. Considering previous observations as well as the results of the present study, it seems that these Fusarium species also follow similar genetic and evolutionary trends.
The estimates of FST and Nm, determined from the IGS data, demonstrated low genetic differentiation and high gene flow among populations. The IGS data showed no genetic distinction between the two Alberta populations (FST = 0, Nm = infinite), while only minor genetic differentiation was found between the Saskatchewan and Alberta populations. The FST value between the Alberta 1998 and Saskatchewan 2000 populations was 0.114, whereas it was 0.088 between the Alberta 2000 and Saskatchewan 2000 populations. The corresponding gene flows (Nm) between these populations were 4.38 and 5.68, respectively. ISSR markers generated from the Alberta 1998 and Saskatchewan 2000 populations also resulted in genetic differentiation and gene flow values (FST = 0.130, Nm = 3.18) similar to those obtained with the IGS data. These results are consistent with a panmictic population genetic structure and further suggest that frequent gene flow and random mating between populations of F. pseudograminearum are predominant evolutionary forces determining the evolution and development of this fungus in western Canada. This explanation is additionally supported by results of the measure of multilocus linkage disequilibrium (rd statistic), which revealed the occurrence of significant recombination in F. pseudograminearum populations. The observed rd value, determined from the IGS data, was 0.2558, which was not significantly different (p = 0.254) from the rd value expected under random mating (rd = 0.2376). Similarly, for the ISSR data, the observed rd value (rd = 0.0031) and the rd value expected under random mating (rd = 0.0029) were also not significantly different (p = 0.427). The occurrence of significant recombination is in agreement with the dynamics of sexual development in the natural populations of F. pseudograminearum [17].
Overall, our study revealed a substantially high genetic diversity within populations of F. pseudograminearum recovered from infected wheat seeds in the provinces of Alberta and Saskatchewan in western Canada. Low genetic differentiation and high gene flow were found among the population samples examined, and significant recombination within populations. These results suggest that regular gene flow and random mating between isolates from different populations could result in new and novel genotypes with both improved pathological and biological traits. As this species has so far been identified in a limited area of the western prairies of Canada, its potential evolution into new, more virulent genotypes is of concern. In fact, both the area affected and the severity of diseases attributable to this pathogen might increase. Thus, there is a need for ongoing monitoring of the incidence and distribution of F. pseudograminearum in Canada. In addition, formulation of an integrated strategy for the management of diseases caused by this fungus deserves attention.
Acknowledgements
A research grant from the Alberta Agricultural Research Institute is acknowledged. We thank Shirley Brezden and Susan Patrick for technical assistance.
References
1. Agapow PM, Burt A (2001) Indices of multilocus linkage disequilibrium. Mol Ecol Notes 1:101-102 [ Links ]
2. Aoki T, O'Donnell K (1999) Morphological and molecular characterization of Fusarium pseudograminearum sp. nov., formerly recognized as the Group 1 of Fusarium graminearum and PCR primers for its identification. Mycologia 91:597-609 [ Links ]
3. Booth C (1971) The genus Fusarium. Commonwealth Mycological Institute, Kew, United Kingdom [ Links ]
4. Burgess LW, Klein TA, Bryden WL, Tobin NF (1987) Head blight of wheat caused by Fusarium graminearum group 1 in New South Wales in 1983. Australasian Plant Pathol 16:72-78 [ Links ]
5. Burgess LW, Backhouse D, Summerell BA, Swan J (2001) Crown rot of wheat. In: Summerell BA, Leslie JF, Backhouse D, Bryden WL, Burgess LW (eds) Fusarium: Paul E. Nelson Memorial Symposium. APS Press, St. Paul, Minnesota, pp 1-294 [ Links ]
6. Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes, application to human mitochondrial DNA restriction data. Genetics 131:479-491 [ Links ]
7. Jiménez-Gasco MM, Navas-Cortés JA, Jiménez-Díaz RM (2004) The Fusarium oxysporum f. sp. ciceris/Cicer arietinum pathosystem: a case study of the evolution of plant-pathogenic fungi into races and pathotypes. Int Microbiol 7:95-104 [ Links ]
8. Laday M, Szecsi A (2002) Identification of Fusarium species by isozyme analysis. Acta Microbiol Immunol Hung 49:321-330 [ Links ]
9. Lee SB, Taylor JW (1990) Isolation of DNA from fungal mycelia and single spores. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols, a guide to methods and applications. Academic Press, New York, pp 283-287 [ Links ]
10. Miedaner T, Schilling AG, Geiger HH (2001) Molecular genetic diversity and variation for aggressiveness in populations of Fusarium graminearum and Fusarium culmorum sampled from wheat fields in different countries. J Phytopathol 149:641-648 [ Links ]
11. Mishra PK, Fox RTV, Culham A (2002) Restriction analysis of PCR amplified nrDNA regions revealed intraspecific variation within populations of Fusarium culmorum. FEMS Microbiol Lett 215:291-296 [ Links ]
12. Mishra PK, Fox RTV, Culham A (2003) Inter-simple sequence repeat and aggressiveness analyses revealed high genetic diversity, recombination and long range dispersal in Fusarium culmorum. Ann Appl Biol 143:291-301 [ Links ]
13. Mishra PK, Tewari JP, Clear RM, Turkington TK (2004) Molecular genetic variation and geographical structuring in Fusarium graminearum. Ann Appl Biol 145:299-307. [ Links ]
14. Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321-3323 [ Links ]
15. O'Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T (2004) Genealogical concordance between mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within Fusarium graminearum clade. Fungal Genet Biol 41:600-623 [ Links ]
16. Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236:787-792 [ Links ]
17. Summerell BA, Burgess LW, Backhouse D, Bullock S, Swan LJ (2001) Natural occurrence of perithecia of Gibberella coronicola on wheat plants with crown rot in Australia. Australasian Plant Pathol 30:353-356 [ Links ]
18. Tóth B, Mesterházy A, Nicholson P, Téren J, Varga J (2004) Mycotoxin production and molecular variability of European and American isolates of Fusarium culmorum. Eur J Plant Pathol 110:587-599 [ Links ]
19. Typas MA, Griffen AM, Bainbridge BW, Heale JB (1992) Restriction fragment length polymorphisms in mitochondrial DNA and ribosomal RNA gene complexes as an aid to the characterization of species and sub-species populations in the genus Verticillium. FEMS Microbiol Lett 95:157-162 [ Links ]
20. Wright S (1951) The genetic structure of populations. Ann Eugen 15:323-354. [ Links ]
21. Zeller KA, Bowden RL, Leslie JF (2003) Diversity of epidemic populations of Gibberella zeae from small quadrats in Kansas and North Dakota. Phytopathology 93:874-880 [ Links ]
![]() Corresponding author
Corresponding author
P.K. Mishra
Department of Molecular
Cellular and Developmental Biology
University of California
Santa Barbara, CA93106-9610, USA.
Tel. +1-805-8933867. Fax +1-805-8934724
E-mail: mishra@lifesci.ucsb.edu
Received 29 November 2005
Accepted 16 January 2006