My SciELO
Services on Demand
Journal
Article
Indicators
-
 Cited by SciELO
Cited by SciELO -
 Access statistics
Access statistics
Related links
-
 Cited by Google
Cited by Google -
 Similars in
SciELO
Similars in
SciELO -
 Similars in Google
Similars in Google
Share
Revista de Osteoporosis y Metabolismo Mineral
On-line version ISSN 2173-2345Print version ISSN 1889-836X
Rev Osteoporos Metab Miner vol.10 n.2 Madrid Apr./Jun. 2018 Epub May 17, 2021
https://dx.doi.org/10.4321/s1889-836x2018000200003
ORIGINALS
Using free access bioinformatic tools to identify potential vascular calcification biomarkers in diabetes mellitus type 2 patients
1Fundación Pública Andaluza para la Investigación Biosanitaria Andalucía Oriental-Alejandro Otero (FIBAO) - Instituto de Investigación Biosanitaria de Granada (Ibs.GRANADA) - Granada (España)
2Unidad de Metabolismo Óseo, UGC Endocrinología y Nutrición - Hospital Universitario Campus de la Salud - Granada (España)
2Instituto de Investigación Biosanitaria de Granada (Ibs.GRANADA) - Granada (España)
3Área temática CIBER de Fragilidad y Envejecimiento Saludable (CIBERFES) - Centro de Investigación Biomédica en Red (CIBER) - Instituto de Salud Carlos III - Madrid (España)
4Servicio de Angiología y Cirugía Vascular - Hospital Universitario Campus de la Salud - Granada (España)
5Servicio de Proteómica - Fundación Pública Andaluza para la Investigación Biosanitaria Andalucía Oriental-Alejandro Otero (FIBAO) - Instituto de Investigación Biosanitaria de Granada (Ibs.GRANADA) - Granada (España)
6Departamento de Medicina - Facultad de Medicina - Universidad de Granada - Granada (España)
Objectives:
Identify potential biomarkers involved in vascular calcification processes to improve DM2 diagnosis and treatment in its subclinical stages.
Methods:
This experimental study included 5 patients suffering diabetes mellitus type 2 (DM2) with peripheral arterial disease and critical ischemia. Protein extraction and identification of the proteome were carried out using liquid chromatography and mass spectrometry (LC-MS/MS) of calcified femoral artery sections. The identified proteins were analyzed through gene ontology and compared with other specific proteins of related vascular pathologies through the DisGeNET database. Cytoscape software analyzed the network of biological functions of the proteins selected for classification based on the disease in which they are involved.
Results:
530 proteins were identified in the analyzed samples with functions mainly of calcium binding and catalytic. 37 of them were common in other related vascular pathologies. The exploration of the biological networks of the 37 proteins identified, led to the identification of 2 potential specific markers of vascular calcification in atherosclerotic processes, such as 10-kDa thermal shock mitochondrial protein, and the flavoprotein subunit of succinate dehydrogenase.
Conclusions:
There is significant expression of proteins involved in processes of bone mineralization in calcified vascular tissue, suggesting the existence of common molecular mechanisms between bone regulation and vascular. The use of bioinformatics tools suggests the involvement of the mitochondrial 10 kDa heat shock protein and the subunit of the succinate dehydrogenase as potential biomarkers of vascular calcification in patients with DM2, although additional studies are needed to confirm this hypothesis.
Introduction
Diabetes mellitus type 2 (DM2) is a significant public health problem throughout the world which affects a large sector of the population. An increased incidence of DM2 cases is predicted worldwide, involving more than 300 million people in 20251. DM2 is associated with various complications, with cardiovascular disorders being one of the main causes of mortality in this disease2. Among the complications, chronic ischemia of lower limbs due to atherosclerotic processes is among the most frequent vascular problems in DM2 patients, with a prevalence of calcified plaques in the lower limbs that may reach 44.6% in this population3. Several studies reveal that DM2 is an independent risk factor in cardiovascular disease (CVD) development in both men and women4,5. The advance of these vascular complications is determined by the presence of common risk factors in DM2 such as obesity, chronic hyperglycemia, insulin resistance, dyslipidemia and states of inflammation and oxidation, among others. However, only some of the patients with DM2 will develop these vascular complications while others, despite presenting the same risk factors, will not suffer cardiovascular events during the disease course. This suggests that the pathway that links dysglycemia and CVD is not well established, requiring in-depth study of the factors involved in the development of CVD associated with DM2. Evidence has shown a process of regulation of vascular calcification similar to bone mineralization, suggesting a connection between bone metabolism disorders and the vascular system.
In this context, the identification of new biomarkers involved in the development of vascular complications in patients with DM2 may be of great importance to improve the prognosis and quality of life of this population, as well as to design new preventive and therapeutic strategies that prevent the development of cardiovascular events, thus reducing the morbidity and mortality of diabetic patients.
Proteomic analysis is a hypothesis-free approach that integrates genetic and epigenetic influences by examining protein expression profiles, and is not limited by prior knowledge. The majority of biomarker search studies have been carried out at the serum level, due to their easy collection by non-invasive techniques. However, there are proteins that are not released into the bloodstream but are expressed specifically in the affected tissues, and therefore cannot be detected by serum proteomic studies. Therefore, the aim of this study is to identify potential biomarkers of CVD in calcified vascular tissue of patients with DM2 through the use of proteomic techniques. One of the main limitations in studies of this type is the obtaining of tissue samples from healthy subjects, or patients without associated complications, which makes it very difficult to carry out a comparative study of the protein profile between cases and controls for the identification of proteins involved in the study.
The introduction of bioinformatics tools in the scientific field in recent years has helped to overcome these limitations. Bioinformatics is crucial to combine information from multiple sources and to generate new knowledge from existing data. It also has the potential to simulate the structure, function and dynamics of molecular systems, among other utilities. Therefore, it may help researchers formulate hypotheses to guide experimental work.
In our study, different bioinformatic tools are applied that allow us to obtain a large amount of information published for free online. In this way, a classifier has been used according to certain characteristics of the proteins identified, such as its molecular function, biological process or its behavior as a cellular component. This allows us to study the proteins as a whole, and their proportion according to these characteristics in the subjects under study.
On the other hand, the use of protein repositories identified as biomarkers of different conditions potentially related to the calcification of the femoral artery allows us to establish protein links between different disorders, as well as to identify specific proteins of this disease.
Therefore, applying these tools allows us to deepen the knowledge of the pathways involved in vascular disorders, offering, at the same time, an experimental orientation aimed at identifying potential biomarkers involved in the atherosclerotic processes of the femoral artery.
Material and methods
Population study
Our experimental work included five subjects. These patients are diagnosed with DM2 according to the criteria of the American Diabetes Association (2011). The criterion for inclusion of peripheral arterial disease was the presence of critical ischemia with indication of a lower limb amputation because they were not candidates for revascularization or because it has failed. Critical ischemia criteria according to recommendation nº 16 of the consensus document on peripheral arterial disease, TASC II [1], are: persistent ischemic pain and relapsing at rest requiring analgesia with opiates for at least 2 weeks; ulcers or gangrene standing or on toes; systolic blood pressure in the ankle less than 50 mmHg or systolic pressure in the toe less than 30 mmHg; chronicity excluding acute ischemia.
The patients came from Granada’s Campus Hospital of Health Studies and were evaluated in the Service of Angiology and Vascular Surgery. All patients were Caucasian, with normal serum levels of calcium and phosphorus and no kidney, liver, gastrointestinal or thyroid diseases. All received medication for DM2, including metformin, sulfonylureas, insulin or a combination of these drugs.
Samples of vascular tissue from the femoral artery with presence of calcification were obtained. The samples were immediately cryopreserved after extraction. All the samples used for the study were managed by the Biobank of the Andalusian Public Health System (SSPA Biobanco) of the San Cecilio University Hospital, in accordance with the Biobank procedures approved by the Ethics Committee for Biomedical Research of Andalusia.
All subjects included in the study signed the acceptance and understanding of informed consent. The study was carried out with the approval of the Ethics Committee of San Cecilio University Hospital and in accordance with the relevant ethical guidelines for human and animal research included in the Helsinki Declaration.
Proteomic study
Protein extraction
Samples corresponding to 45 sections of femoral artery tissue were homogenized at 4°C after the addition of 500 µL of soluble protein extraction buffer (20 mM Tris-HCl pH 7.6, 10 mM NaCl, 0.5 mM deoxycholate sodium, 1 mM EDTA, 4% SDS, 30% glycerol, 5 mM PMSF, 200 mM DTT, benzonase -1µL/10 mL of buffer-). The samples were centrifuged for 15 minutes at 6,000 r.p.m. and 4°C and the supernatant representing the cytosolic fraction of the cells where the soluble proteins are found was collected. These samples were stored at -80ºC until further processing for protein analysis.
Sample processing
The frozen samples were sent for analysis to the Proteomics Unit of the University of Córdoba’s Central Research Support Services (SCAI). Once thawed, the samples were concentrated by ultrafiltration using an Amicon Ultra-0.5 Centrifugal Filter spin column of 3 kDa. Protein extracts were cleaned by SDS-PAGE 1D 10% polyacrylamide electrophoresis. For this, the samples were loaded on the concentrator gel, to which a voltage of 100 V was applied until the electrophoresis front reached the separating gel. The assay was stopped when the protein extract had entered 1 cm into the separating gel. The gel was stained with Coomassie blue, and then the cutting and extraction of the gel bands that were kept in water until digestion was carried out.
For protein digestion, the gel portions were preserved in 50 mM ammonium bicarbonate (BA)/50% acetonitrile for 15 minutes, with subsequent incubation for 5 minutes in 100% acetonitrile. The protein was maintained under reducing conditions by the addition of 20 mM DTT in 25 mM BA, and was incubated for 20 minutes at 55°C. The mixture was cooled to room temperature, with the subsequent alkylation of the free thiol groups by the addition of 40 mM iodoacetamide in 25 mM BA for 20 minutes in the dark. Subsequently, the gel portions were washed twice in 25 mM BA. Proteolytic digestion was carried out by the addition of trypsin (Promega, Madison, Wisconsin, USA) to 12.5 ng/µL of enzyme in 25 mM BA, and incubated at 37°C overnight. The digestion of proteins was stopped by adding trifluoroacetic acid to a final concentration of 1%. The digested samples were finally dried by SpeedVac.
Analysis by Mass Spectrometry/Mass coupled with liquid nano-Chromatography (nLC-MS/MS)
The Nano-LC was carried out on a UPLC Dionex Ultimate 3000 nano (Thermo Scientific) with an Acclaim PepMap100, C18, 3 µm, 100 Å, 75 µm i.d. x 50 cm, nanoViper (Thermo Scientific). The peptide mixture was preloaded in a 300 µm x 5 mm Acclaim Pepmap precolumn (Thermo Scientific) in 2% acetonitrile/0.05% trifluoroacetic for 5 min at a flow of 5 µL/min. Peptide separation was carried out at 40°C for all runs. The elution buffers were: buffer A (0.1% formic acid in water) and buffer B (20% acetonitrile and 0.1% formic acid). Samples were eluted at a flow of 300 nL/min with the following gradient: 4-35% B for 120 minutes; 35-55% of B for 6 minutes; 55-90% of B for 3 minutes, followed by 8 minutes of washing with 90% B and reequilibration from 15 minutes to 4% of B. The total time of the chromatography was 150 minutes.
The eluted peptide cations were converted to gas phase ions by nanoelectrospray ionization and analyzed in a mass spectrometer, Thermo Orbitrap Fusion (Q-OT-qIT, Thermo Scientific) operated in positive mode. The recognition scan of the peptide precursors from 400 to 1500 m/z was carried out at 120,000 FWHM resolution with the objective of counting 4 x 105 ions. Tandem mass spectrometry was carried out by isolation at 1.2 Da with the quadrupole, CID fragmentation with normalized collision energy of 35 and fast scanning MS analysis in the ion trap. The AGC ion count target was adjusted to 2 x 103 and the maximum injection time was 300 ms. Only those precursors with loading status of 2-5 were sampled for MS/MS. The dynamic exclusion duration was set at 15 s with a tolerance of 10 ppm around the selected precursor and its isotopes and activating the selection of monoisotopic precursors.
Identification of proteins
The raw data was processed using the Proteome Discoverer software package (version 2.1.0.81, Thermo Scientific). MS/MS spectra were searched with the SEQUESTTM6 engine (Thermo Fisher Scientific) against a Uniprot_Homosapiens_ dateofanalisys database (www.uniprot.org). A theoretical tryptic digestion of the peptides was carried out, cysteine carbamidomethylation was introduced as a fixed post-translational modification and methionine oxidation as a variable post-translational modification. The tolerance of the mass of the precursors was 10 ppm and the ions of the product were recorded with a tolerance of 0.1 Da. The spectral matches of the peptides were validated using a filter based on q values at 1% FDR (False Discovery Rate), obtaining a list of proteins identified in each of the processed samples (Proteome Discoverer).
Data analysis using bioinformatics tools
Analysis by gene ontology (GO)
The gene ontology resource (gene ontology) is an important initiative of bioinformatics that aims to standardize the representation of protein attributes between species and databases7. The online system provides a vocabulary of terms to describe characteristics of the gene products and annotation data of gene products of the members of the GO consortium, as well as tools to enter and process this data. Each protein is characterized in terms of three ontologies: molecular function, cellular component and the biological process involved8. Using the GO database (http://www.geneontology.org) and the Onto-Express analysis, the genes involved were classified in order to obtain an overview of the potential functions of the expressed genes corresponding to the proteins identified. in the study.
Analysis by comparison of pathologies
The proteins identified in this study were confronted with the proteins already described in other vascular pathologies with a possible relationship with the vascular calcification of the femoral artery. This information was obtained thanks to the DisGeNET database (http://www.disgenet.org) that contains one of the largest collections of genes and variants associated with human diseases, publicly available9. The pathologies from which the information was obtained were: Carotid Stenosis, UMLS CUI: C0007282; Supravalvular aortic Stenosis, UMLS CUI: C0003499; Aortic Valve Stenosis, UMLS CUI: C0003507; Esophageal Stenosis, UMLS CUI: C0014866; Laryngostenosis, UMLS CUI: C0023075; Mitral Valve Stenosis, UMLS CUI: C0026269; Pulmonary Valve Stenosis, UMLS CUI: C0034194; Pyloric Stenosis, UMLS CUI: C0034194; Aortic Calcification, UMLS CUI: C1096249; Calcification of Mitral Valve, UMLS CUI: C0919718; Aortic Valve Calcification, UMLS CUI: C0428791; Renal Artery Stenosis, UMLS CUI: C0035067 and Vascular Calcification, UMLS CUI: C0342649. The visualization of these proteins was carried out using the Cytoscape tool10.
Analysis by protein-protein interaction and functional networks
To investigate the direct (physical) and/or indirect (functional) relationships between identified genes, we used the search tool for the retrieval of interacting genes (STRING) in databases to analyze the functional network (https://string-db.org) . The STRING database provided a score for each gene-gene interaction, calculated as the joint probability of the different evidence channels (protein interaction, fusion, coexpression, etc.), correcting the random probability of observing an interaction. A high database score meant that there was high experimental or predicted evidence for functional gene-gene interaction. Therefore, a functional associated network was constructed on the basis of the expression profile of the proteins identified in the present study.
Results
Gene ontology
We identified 530 proteins from the processed samples (p<0.05, q value=0). All the identified proteins were classified using the gene ontology annotations and grouped into the three functional groups: biological processes, cellular components and molecular functions (Figure 1).
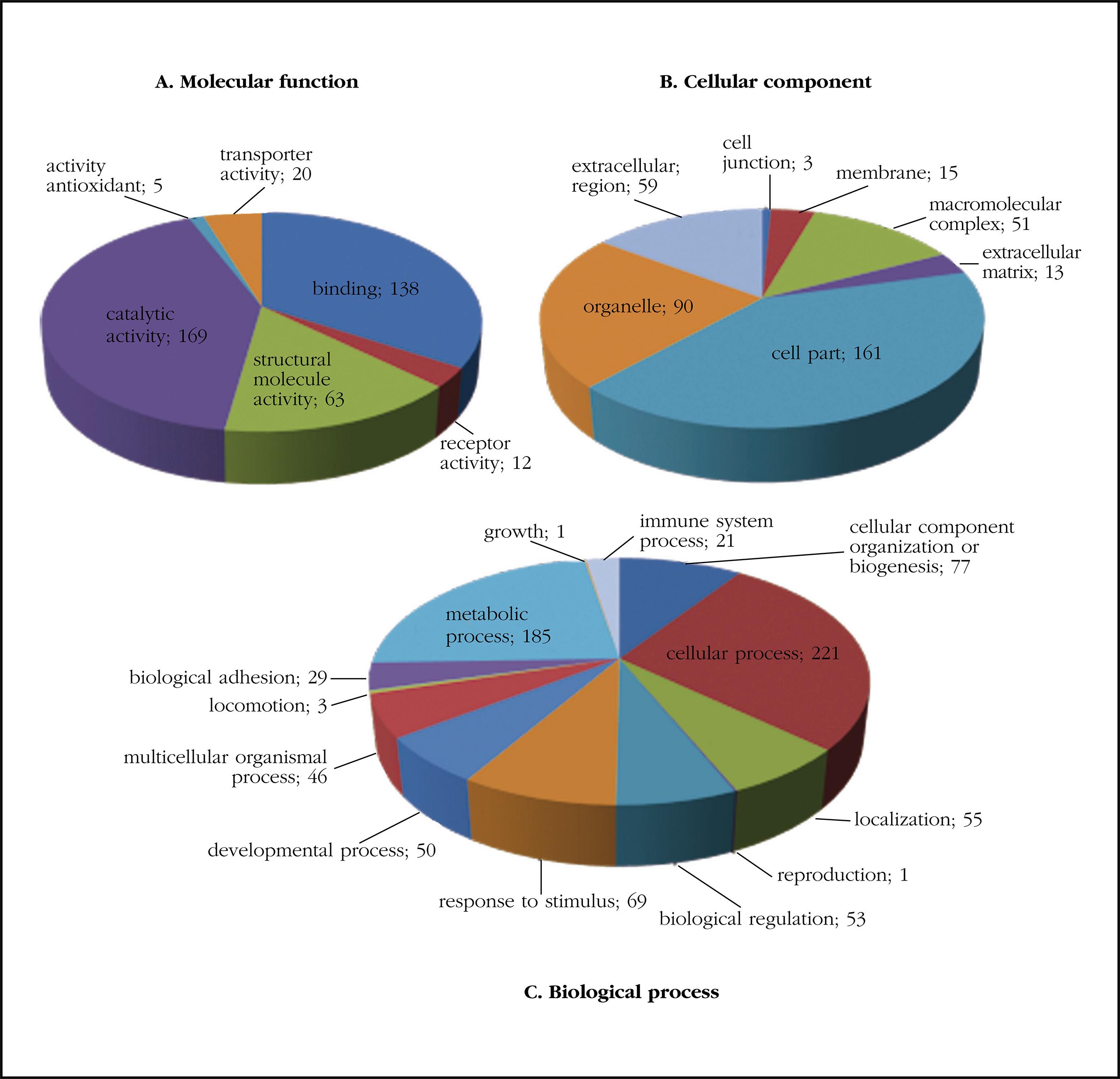
Figure 1. Protein classification based on gene ontology according to molecular functions (A), cellular component (B) and biological process (C)
The analysis of the gene ontology showed that the two most common molecular functions in the selection carried out were functions related to catalytic activity (GO: 0003824) and with binding (GO: 0005488). Regarding the analysis of the cellular component, the most frequent was the cellular portion (GO: 0044464), followed by organelle (GO: 0043226) and extracellular region (GO: 0005576); as biological processes highlighted those related to cellular processes (GO: 0009987), metabolic processes (GO: 0008152) and organization or biogenesis of cellular components (GO: 0071840).
Table 1 shows the main classification according to GO of the proteins that have been identified classified in protein classes. It should be noted a high proportion of calcium binding proteins.
Protein comparison
Of the 530 proteins identified in the proteomic study, 37 proteins coincided with proteins already described in vascular pathologies, some of which were recurrent in different vascular pathologies (Table 1). These proteins were selected as candidates to perform a study of their biological network in order to know their relationship with vascular calcification in the femoral artery (Figure 2).
Functional networks
The 37 proteins of interest resulted in an extensive functional network where a nucleus representing the calcification of the femoral artery is observed, where numerous proteins that are also part of other related diseases converge (Figure 3).
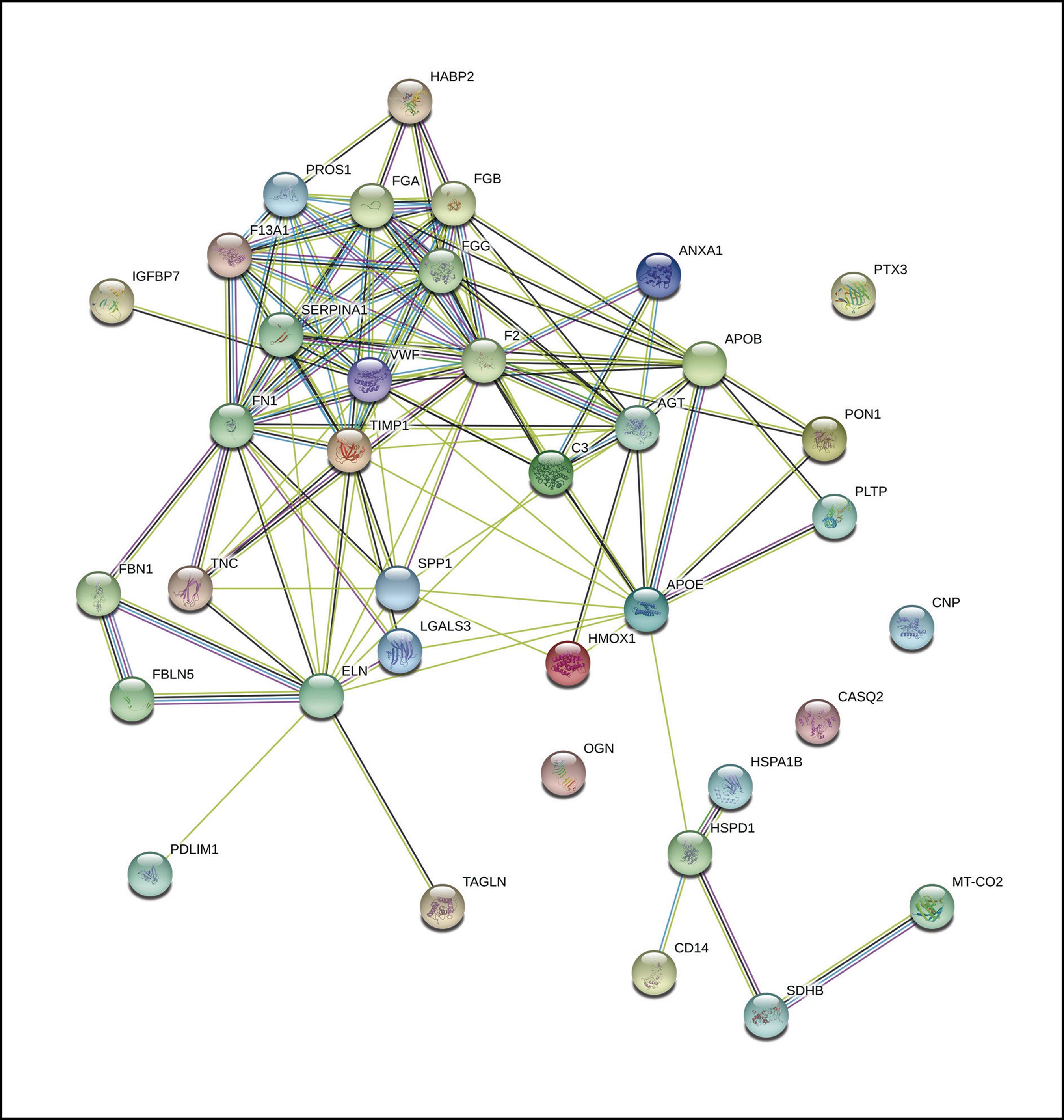
Figure 3. Functional network of the 37 proteins identified by nLC-MS/MS in calcified femoral artery samples from patients with DM2, previously described in vascular diseases
To this first functional network, 5 more proteins were added related to the initial network (Figure 4). The addition of this second level allowed us to find two of the five proteins included in the network as possible specific biomarkers of femoral artery calcification, as they were not found in the rest of the related diseases studied.
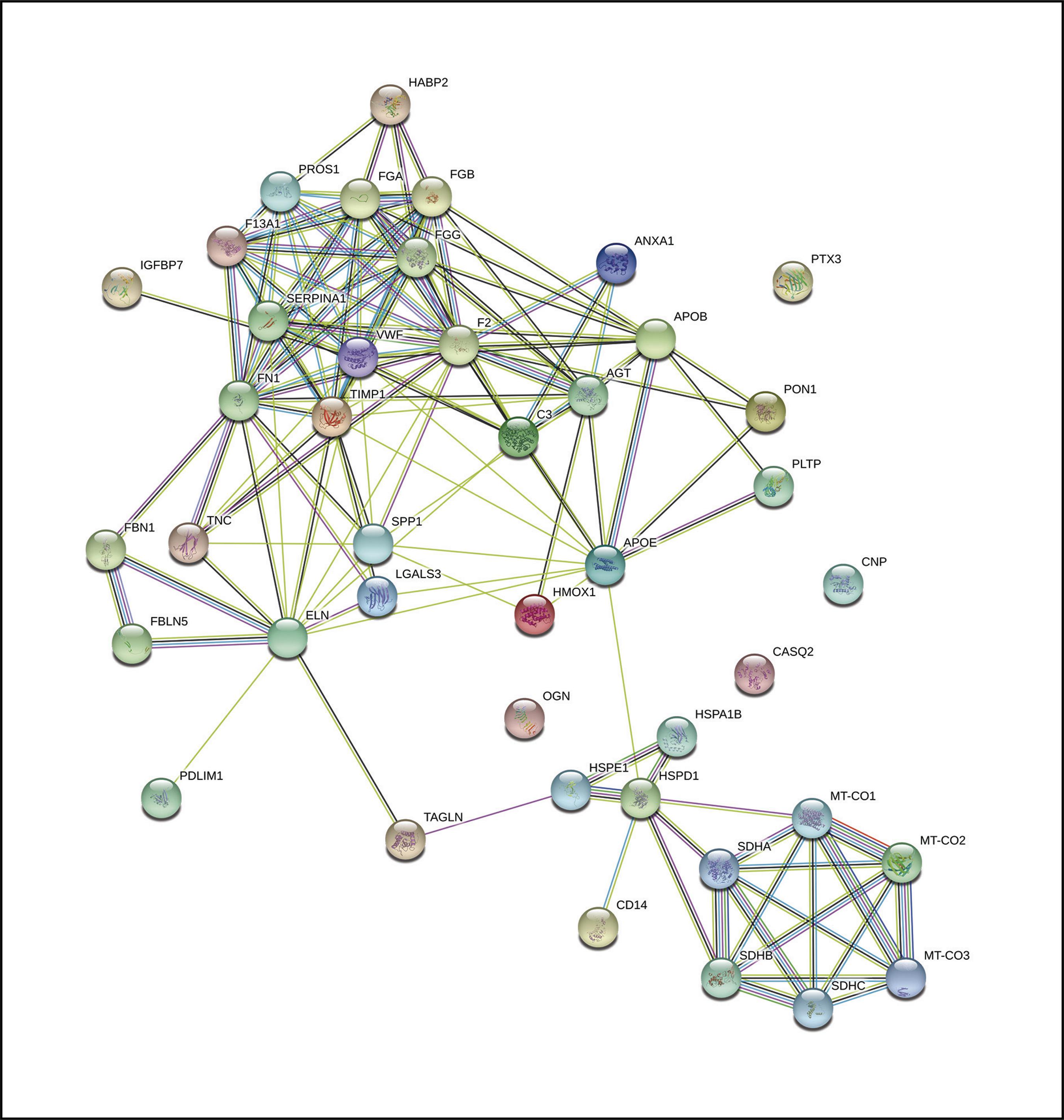
Figure 4. Functional network of the 37 proteins identified by nLC-MS/MS in calcified femoral artery samples from patients with DM2, previously described in vascular disorders, after the addition of 5 new related proteins
These two proteins were also identified in our proteomic analysis (flavoprotein subunit of succinate dehydrogenase (SDHA, P31040), mitochondrial 10 kDa heat shock protein (HSP1, P61604), expressed in samples from patients with femoral artery calcification Therefore, they were selected as the first two candidates to begin their study in greater depth.
Discussion
DM2 currently represents a serious health problem as it affects a large proportion of the population. Patients with DM2 have a higher risk of developing various complications, mainly cardiovascular events that represent the main cause of mortality of this disease. The probability of developing vascular complications in this population depends on a series of traditional factors and others that are not completely known. The identification of new factors involved in these disorders can facilitate the early diagnosis of high-risk subjects before irreversible damage occurs. The aim of the present study was to identify proteins in vascular tissue that could be related to processes of vascular calcification and, therefore, to increased cardiovascular risk in subjects with DM2.
Studies into biomarker identification at the proteomic level present a high complexity due to the large amount of proteins and pathways involved in the different diseases, the interaction between proteins, protein differences depending on the study tissue, high technological costs, among others. All this, in addition to the large amount of data to be processed that are generated in these studies, makes it difficult, in many cases, to identify real biomarkers of certain diseases. To all this we must add the difficulty of obtaining tissue samples from control subjects in order to carry out a comparative study between the different study groups.
The use of bioinformatics tools allows, to a large extent, to solve many of the problems that arise in biomarker identification studies. The use of these tools facilitates the study, comparison and correlation of data found experimentally; the prediction of domains, structures and phylogenetic relationships of the sequences examined; the development of algorithms and statistics necessary for the understanding of biological information, interconnecting information from different approaches. The information provided by these tools allows, in addition, an orientation of the scientific work offering keys that allow a saving of time and economic and human resources.
Today, bioinformatics tools, available for free on the net and increasingly efficient for in silico studies, are booming.
This study establishes a simple and intuitive method to locate biomarkers related to vascular calcification processes. After the identification of 530 proteins corresponding to the characteristic femoral artery protein profile with presence of calcification of patients with DM2 using nLC-MS/MS, various bioinformatic strategies have been used to limit and filter the search for biomarkers, as well as discard those that do not were related to vascular pathologies. In the first place, the gene ontology methodology was used, which focuses on the study of the functions of the group of genes corresponding to the identified proteins, in order to select those functions most related to processes involved in vascular calcification. The majority molecular function of the genes coding for the identified proteins was the catalytic function and the binding function. Recent studies have shown that, in a calcifying environment, vascular smooth muscle cells are able to undergo a phenotypic transition to osteocyte-like cells capable of expressing typical bone markers, contributing in this way to arterial mineralization12. It is not surprising that, under these conditions, there is an increase in the proteins that bind calcium, thus contributing to the calcium deposition in the arterial wall. These processes lead to the development of atherosclerotic processes so frequent in patients with DM2. Recent studies show that macrophages surrounding calcium deposits in human atherosclerotic plaques are phenotypically defective and incapable of reabsorbing calcification13. In this line, the identification of proteins with catalytic activity in the vascular samples analyzed, with the presence of calcifications, could reflect a defense mechanism of the organism to try to disintegrate the components of the atheroma plaque generated in these situations. According to this hypothesis, there are studies that show an increase in markers of bone resorption in patients with vascular disorders14,15.
With the aim of directing and delimiting the biomarker identification study, a comparison was made of the 530 proteins identified in the proteome of the study subjects with those described as proteins involved in various vascular diseases, in the repository of molecular markers (DisGeNet). By using this tool, the number of candidate proteins was reduced to 37, those commonly expressed in various vascular disorders associated with vascular calcification processes. The disposition of these protein nexuses in a biological network shows a potential capacity as a biomarker of some of the proteins, which participate in various vascular conditions. The design of specific interaction networks between each of the 37 selected proteins showed a well-formed interaction network, in which a close relationship is observed between most of these proteins. Among these proteins, we find some that could be indirectly related to calcification processes, such as osteopontin and osteoglycine, both of which are involved in biomineralization and bone remodeling processes. Likewise, we also identify muscle proteins typical of vascular smooth muscle cells such as transgelin or tropoelastin, which could indicate the loss of the vascular phenotype. The presence of other proteins of interest identified, but which did not pass the level of statistical significance, as is the case of osteoprotegerin or the GLA protein of the matrix, could be due to the low number of study subjects, which generates a variability that is high enough to be statistically discarded. However, by increasing the interaction network by 5 proteins, adding the first five proteins closely linked to those arranged in the first network, we observed that 2 of them are part of the proteome characteristic of the vascular tissue samples analyzed. One of them, the flavoprotein subunit of succinate dehydrogenase (P31040), is involved in several processes, such as the synthesis of fumarate to succinate in the tricarboxylic acid cycle of carbohydrate metabolism, or in oxide-reduction processes, among others. The deficiency of succinate dehydrogenase is related to a lower flow of electrons and a decrease in antioxidant activity in the respiratory chain, increasing oxidative stress and hypoxia, with important consequences in the process of vascular calcification.
The presence of this protein in calcified vascular tissue could be related to a defensive mechanism to preserve the production of aerobic energy in the state of hypoxia generated in the process of vascular calcification during atherosclerosis16. The other, a 10 kDa, mitochondrial heat shock protein (HSPE1, P61604), is a co-chaperone involved in the importation of mitochondrial proteins and macromolecular assembly. In addition, it is also involved in osteoblastic differentiation and in the activation of endopeptidase activity during the apoptotic process.
The fact that these two proteins do not appear in the rest of the related pathologies that were compared in the study, suggests that they could be two good candidates to suggest them as specific biomarkers of atherosclerosis in the lower limbs in DM2.
Our study presents certain limitations. First, the limited number of control subjects makes comparison with a reference study group impossible. Secondly, the small number of study subjects leads to a high variability of the results, limiting the identification of potential proteins of interest.
Although future studies are needed to confirm the role of the proteins identified as possible biomarkers of vascular calcification in diabetic patients, the use of bioinformatics tools has allowed us to generate hypotheses to work on thanks to the simplification of the large amount of data obtained. The identification of proteins with a high degree of expression in calcified vascular tissue in patients with high cardiovascular risk could offer information to achieve an early and targeted treatment.
Funding:The present work has been financed through a research grant from the FSEEN in the 2016 call and from a project of the Junta de Andalucía (PI0207-2016).
REFERENCES
1. Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307(5708):384-7. [ Links ]
2. Jensen MK, Bertoia ML, Cahill LE, Agarwal I, Rimm EB, Mukamal KJ. Novel metabolic biomarkers of cardiovascular disease. Nat Rev Endocrinol. 2014; 10(11):659-72. [ Links ]
3. He C, Yang J, Li Y, Rong J, Du F, Yang Z, et al. Comparison of lower extremity atherosclerosis in diabetic and non-diabetic patients using multidetector computed tomography. BMC Cardiovasc Disord. 2014;14:125. [ Links ]
4. Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97(18):1837-47. [ Links ]
5. Wilson PW. Diabetes mellitus and coronary heart disease. Am J Kidney Dis. 1998;32(5 Suppl 3):S89-100. [ Links ]
6. Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976-89. [ Links ]
7. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25-9. [ Links ]
8. Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258-261. [ Links ]
9. Piñero J, Queralt-Rosinach N, Bravo À, Deu-Pons J, Bauer-Mehren A, Baron M, et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database J Biol Databases Curation. 2015;2015:bav028. [ Links ]
10. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-504. [ Links ]
11. von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33:D433-7. [ Links ]
12. Zhu D, Mackenzie NCW, Millán JL, Farquharson C, MacRae VE. The appearance and modulation of osteocyte marker expression during calcification of vascular smooth muscle cells. PloS One. 2011;6(5):e19595. [ Links ]
13. Chinetti-Gbaguidi G, Daoudi M, Rosa M, Vinod M, Louvet L, Copin C, et al. Human alternative macrophages populate calcified areas of atherosclerotic lesions and display impaired RANKL-induced osteoclastic bone resorption activity. Circ Res. 2017;121(1):19-30. [ Links ]
14. Morisawa T, Nakagomi A, Kohashi K, Kusama Y, Shimizu W. Serum Tartrate-resistant acid phosphatase-5b levels are associated with the severity and extent of coronary atherosclerosis in patients with coronary artery disease. J Atheroscler Thromb. 2017; 24(10):1058-68. [ Links ]
15. Pasqualini L, Ministrini S, Macura A, Marini E, Leli C, Siepi D, et al. Increased bone resorption: a possible pathophysiological link between hypovitaminosis D and peripheral arterial disease. Eur J Vasc Endovasc Surg. 2016;52(3):352-9. [ Links ]
16. Lukyanova LD, Kirova YI. Mitochondria-controlled signaling mechanisms of brain protection in hypoxia. Front Neurosci. 2015;9:320. [ Links ]
Received: November 23, 2017; Accepted: January 28, 2018











 text in
text in 




