INTRODUCTION
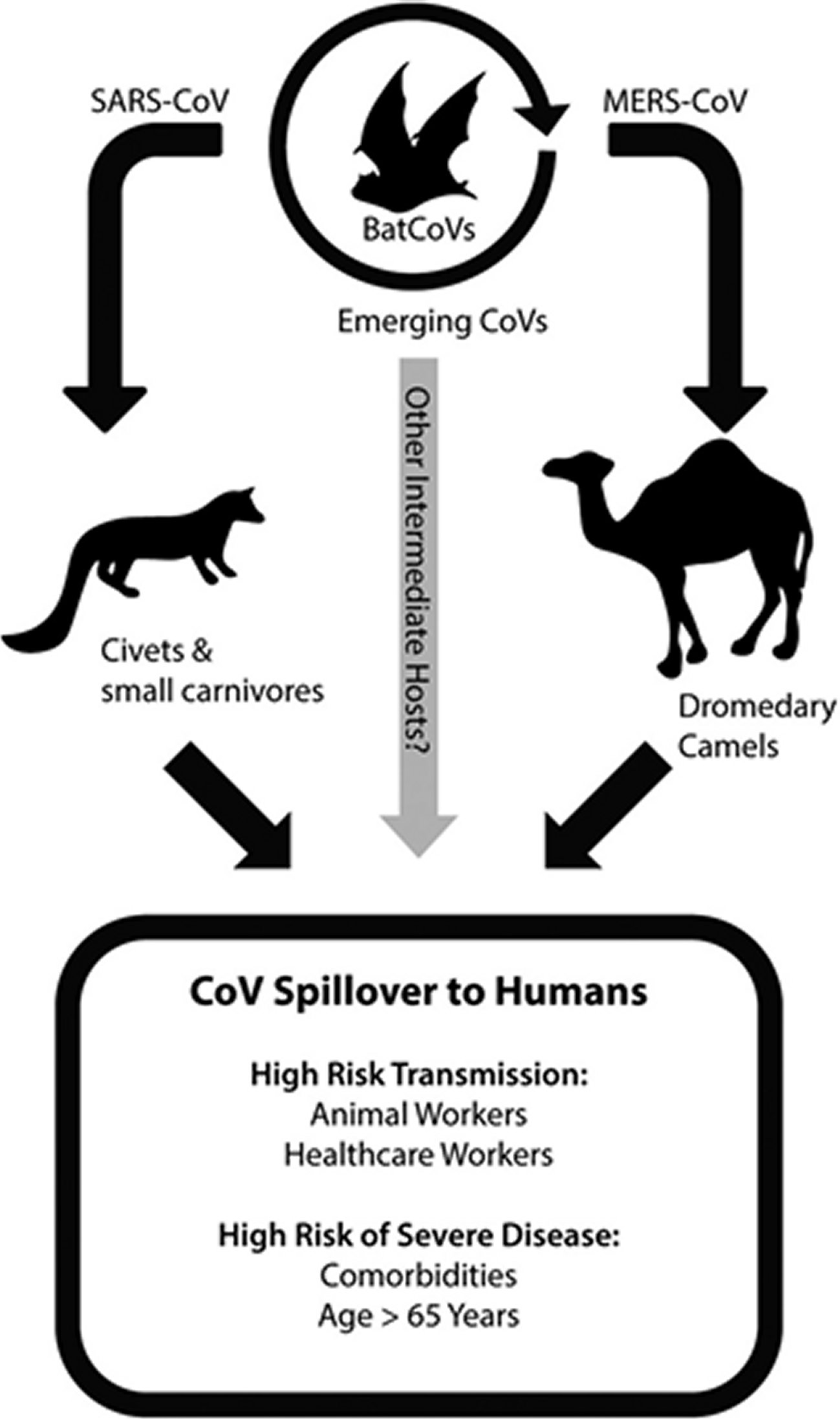
Currently a worldwide pandemic of a new type of Coronavirus (COVID-19) is going on, that originated from Wuhan, China and has now spread to 140 other countries including Japan, Korea, Italy, USA and UK. The World Health Organization (WHO) declared that COVID-19 has assumed pandemic proportions, causing severe respiratory tract infections (RTI) in humans. Present evidence points that SARS-CoV2 spreads to humans via transmission from wild animals illegally sold in the Huanan Seafood Wholesale market. Phylogenetic analysis demonstrated that SARS-CoV2 is a new member of the Corona viridae family, but is separate from SARS-CoV (identity of about 79%) as well as MERS-CoV (identity of about 50%) [1, 2, 3]. This SARS-CoV2 shares a high level genetic similarity (96.3%) with the bat Coronavirus RATG13, that was received from bats in Yunnan in 2013, but the bats are not the immediate source of SARS-CoV2 [4]. Bats have been identified as the natural reservoirs as well as vectors of a lot of coronaviruses; with these viruses having crossed species barriers to infect humans as well as many various types of animals, including avians, rodents and chiropters (Figure 1) [5].
Courtesy of reference number 5
Figure 1. Coronavirus emerges from zoonotic reservoirs.Emergence of coronaviruses into human populations, including highly pathogenic viruses like SARS-CoV and MERS-CoV, has occurred by spillover from bat reservoir hosts into intermediate hosts. The intermediate hosts during the 2003 SARS-CoV epidemic included civets and other small carnivore species located in wet animal markets. MERS-CoV has been identified in dromedary camels, and is particularly associated with active infection of juvenile camels. Novel emerging CoVs may occur in the future via infection from bat populations into other intermediate animal hosts. Additional evidence from BatCoVs indicates that pre-emergent CoVs with the ability to directly infect human cells may be poised for emergence into human populations. Based on prior research from SARS and MERS outbreaks, animal workers that have contact with intermediate animal host species and health-care workers that are exposed to nosocomial CoV infections are at increased risk of highly pathogenic coronavirus transmission. More severe disease in SARS and MERS cases resulted in patients that were over the age of 65 or had comorbidities such as obesity, heart disease, diabetes, renal disease, or hypertension.
EPIDEMIOLOGY
A study of the early transmission dynamics of COVID-19 showed that the mean incubation period was 5.2 days (95% confidence interval [CI], 4.1-7.0) with the 95% percentile of the distribution at 12.5 days [6]. A later study utilizing the travel history and symptom onset of 88 confirmed cases demonstrated a similar mean incubation period of 6.4 days (95% confidence interval [CI], 5.6-7.7) [7]. An unusual case was also documented where incubation period was as long as 19 days [8]. The 19 day incubation period is a low probability event and experts point that a 14 days of quarantine is required usually.
STRUCTURE OF SARS-COV ALONG WITH SARS-COV-2
Coronaviruses represent a family of large enveloped, positive stranded RNA viruses which cause upper respiratory, gastrointestinal tract (GIT), as well as central nervous system (CNS) diseases in humans as well as other animals [9]. Human Coronaviruses H CoV-OC43. H CoV-229E, H CoV-NL63 as well as H CoV-HKU1 circulate in humans and cause mild respiratory diseases [10]. But the outbreak of SARS-CoV2 in 2002 as well as MERS-CoV in 2012 demonstrated that Coronaviruses can cross the species barrier and come out as highly pathogenic viruses [11]. The high mortality rate along with wide spread nature of these new emerging Coronaviruses point that they are a marked threat to global health.
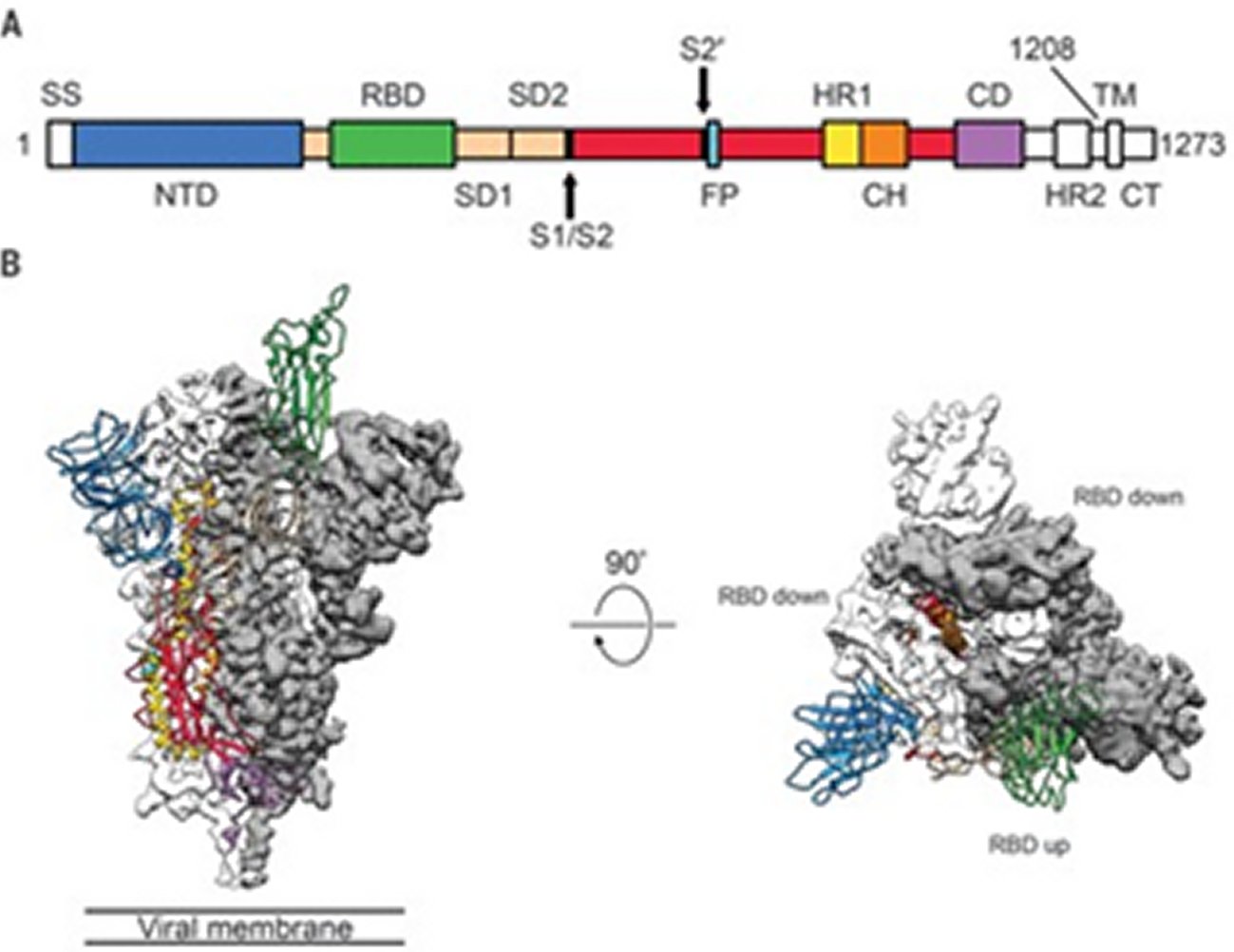
The spike(S) glyco protein of the Corona viruses is a class I viral fusion protein present on the outer envelope of the virion having a key role in viral infection by recognizing host cell receptors and modulating fusion of the viral as well as cellular membranes [12]. The Coronavirus S glyco protein gets synthesized as a precursor protein made up of ~1300 amino acids which then gets cleaved into an amino(N)terminal S1 subunit (~700 amino acids) as well as carboxy(C) terminal S2 subunit(600 amino acids). 3 S1/S2 heterodimers assemble to develop a trimer spike that protrudes via the envelope. The S1 subunit contains a receptor binding domain (RBD), while the S2 subunit possesses a hydrophobic fusion peptide as well as 2 heptad repeat regions. Triggered by receptor binding, proteolytic processing as well as acidic pH in the cellular compartments, the class 1 viral fusion proteins undergo a transition through a metastable prefusion state to a stable prefusion state at the time of infection. Receptor binding subunit gets cleaved as well as the fusion subunits undergoes large conformational rearrangements for exposure of the hydrophobic fusion peptide, stimulating the development of a 6-helix bundle, and bring the viral along with cellular membranes nearer for fusion [13].
Coronavirus S glyco protein has 2 separate protease cleavage sites. The S1/S2 cleavage site of the SARS-CoV S glyco protein is present following residue 667 of the precursor protein, while the S2’ cleavage site of the SARS-CoV S glyco protein is on the S2 subunit and is 130 amino acids from the N terminus of the S2 subunit [14]. The S1/S2 cleavage site is present in a flexible loop of residues 660-675, which is totally exposed in the prefusion S1-S2 trimer spike. The S2’ cleavage site of the SARS-CoV S glyco protein is markedly conserved among Coronaviruses as well as is totally bound in the prefusion SARS-CoV S glyco protein [14]. Cleavage of the S2’ site by host cell proteases is needed for successful infection by SARS-CoV [15]. But the mechanisms involved in the exposure as well as cleavage of the S2’ cleavage site are not clear. Structural biology studies especially recent cryo-electron microscopy (cryo-EM) studies have furthered us getting insight in the role of Corona virus S glyco protein in virus entry. S glyco protein structures in the prefusion state have been documented regarding members of Alpha Coronavirus genus (H CoV-NL63), Beta Corona virus genus (mouse hepatitis virus (MHV), HKU1, SARS-CoV along with MERS-CoV), Gamma Coronavirus genus (IBV) as well as Delta Corona virus genus (Pd CoV) [14, 16, 17, 18, 19, 20, 21]. Prefusion S glyco protein takes up a similar mushroom like homo-trimer architecture, of which the stem is basically made up of 3 S2 subunits, with the top cap made of 3 interwoven S1 subunits. A recently reported cryo EM structure of the MHV S glyco protein in the postfusion state demonstrates an elongated cone-shaped structure which is significantly separate from the prefusion structure and points that marked conformational changes take place during the prefusion to postfusion transition [22] (Figures 2, 3 and 4).
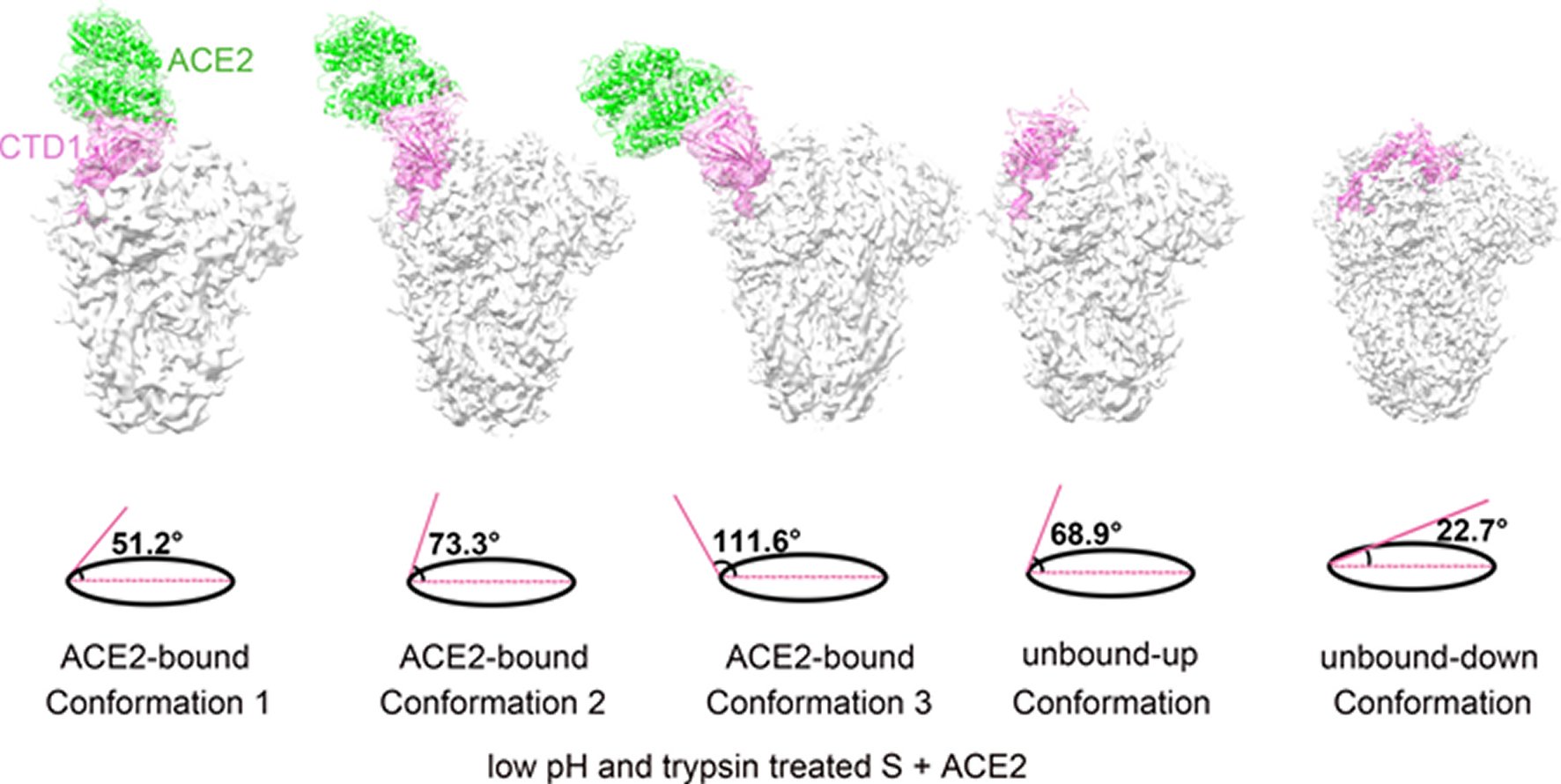
Figure 2. GS-441524 and GS-5734 inhibit MHV with minimal cytotoxicity.(A) GS-441524 is a 1′-cyano 4-aza-7,9-dideazaadenosine C-adenosine nucleoside analogue. (B) Change in viral titer of MHV compared to vehicle control after treatment with GS-441524. The data represent the results from 2 independent experiments, each with 3 replicates. Error bars represent standard error of the mean (SEM). (C) Viral titer data from panel B presented as the percentage of uninhibited control. The EC 50 of GS-441524 was calculated to be 1.1 μM. (D) Cell viability normalized to the vehicle control after treatment with GS-441524. The data represent the results from 3 independent experiments, each with 3 replicates. Error bars represent SEM. (E) GS-5734 is a monophosphoramidate prodrug of GS-441524. (F) Change in viral titer of MHV compared to vehicle control after treatment with GS-5734. The data represent the results from 4 independent experiments, each with 3 replicates. Error bars represent SEM. (G) Viral titer data from panel F presented as the percentage of uninhibited control. The EC 50 of GS-5734 was calculated to be 0.03 μM. (H) Cell viability normalized to vehicle control after treatment with GS-5734. The data represent the results from 3 independent experiments, each with 3 replicates. Error bars represent SEM.
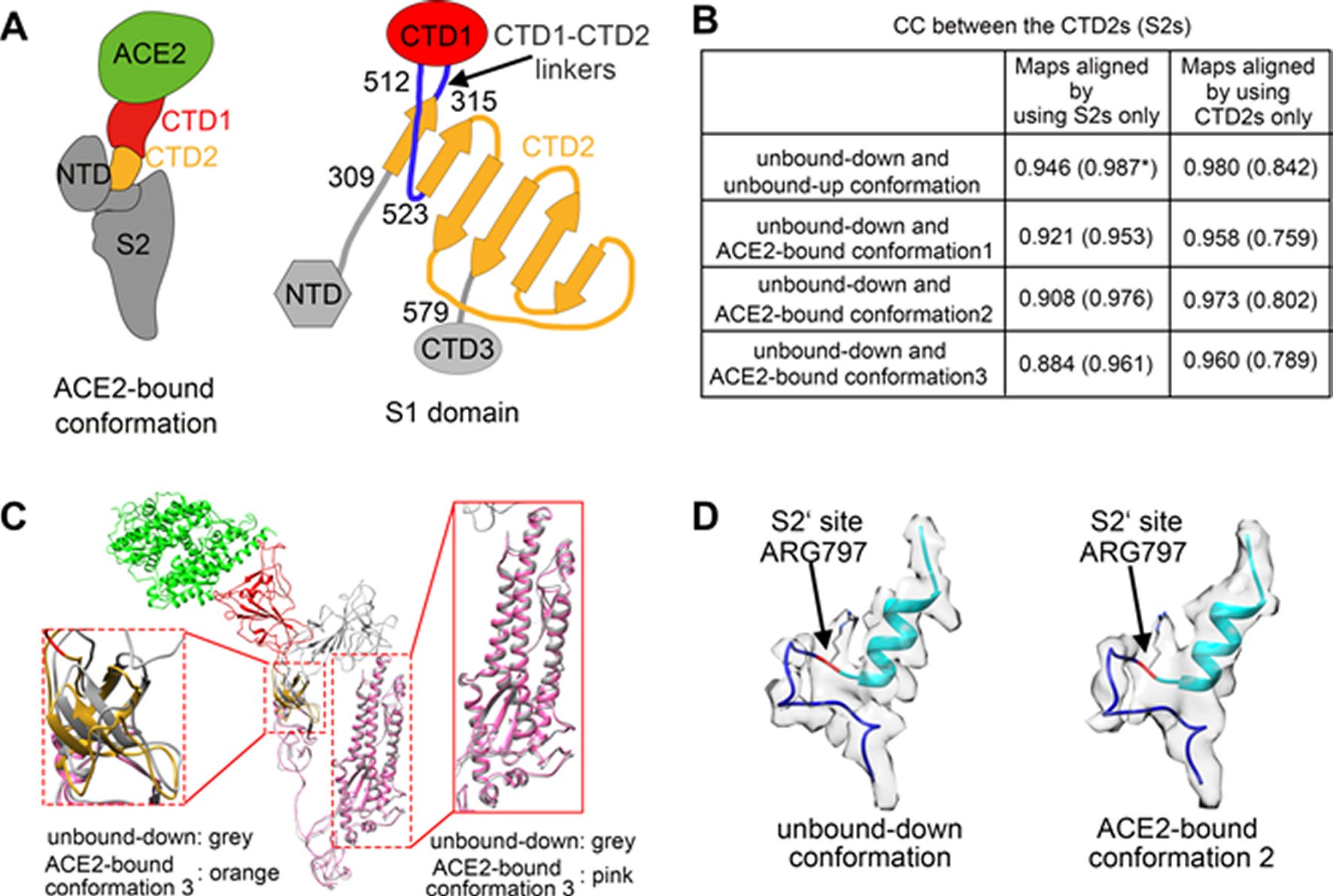
Figure 3. Structural comparisons of the ACE2-bound and ACE2-free SARS-CoV spikes.(A) Schematic and topology diagrams showing the domain organization. NTD, CTD3 and S2 are colored grey, CTD1 is colored red, CTD2 is colored orange, CTD1-CTD2 linkers are colored blue. (B) Cross-correlation coefficients (CCs) between the CTD2s or the S2s of different conformations. *Values in the parentheses are the CCs between the S2s of different conformations. Density maps were low-pass filtered to 5.5 Å and were compared at a contouring level of 4.0 σ. (C) Ribbon-diagram structural comparisons of the ACE2-bound conformation 3 and the unbound-down conformation. ACE2, CTD1, CTD2, CTD3 and S2 of the ACE2-bound conformation 3 are colored green, red, yellow, pink and pink respectively. The unbound-down conformation is colored grey. CTD2 and S2 domain are zoomed in to show the receptor-binding induced hinge motion of CTD2. (D) EM densities and corresponding atomic models represented in ribbon diagrams around the S2’ protease cleavage site: unbound-down conformation (left) and ACE2-bound conformation (right). The S2’ site is colored red and position of the S2’ site is indicated with black arrows. The fusion peptide is colored cyan. The “C” shape loop covering the S2’ site is colored blue.
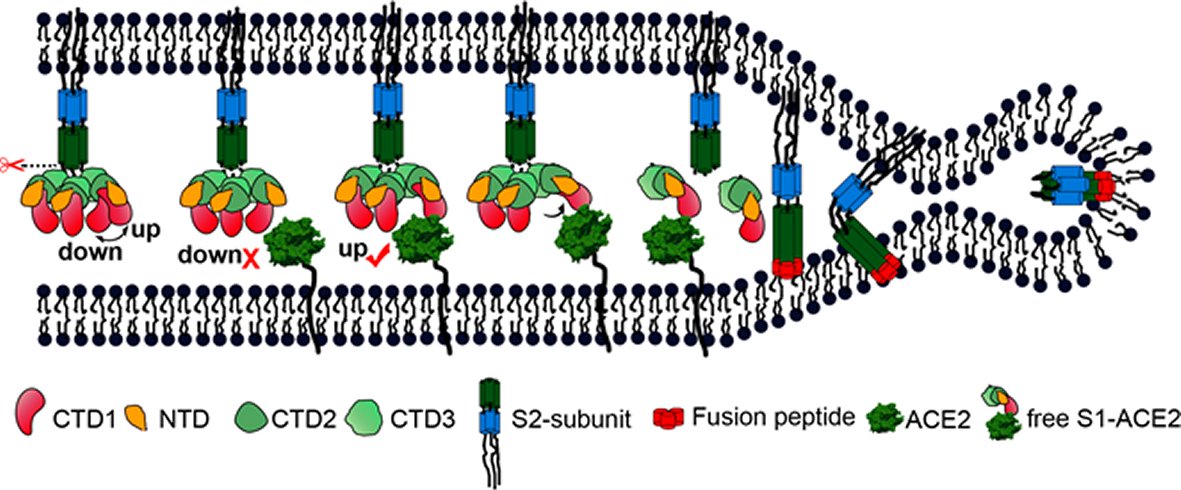
Figure 4. A cartoon representation showing the pre- to post-fusion transition of the SARS-CoV S glycoprotein.The “down” to “up” transition of the receptor-binding domain (CTD1) allows receptor binding. The binding to ACE2 opens up CTD1 and CTD2, promotes the disassociation of the S1-ACE2 complex from the S1/S2 cleaved S glycoprotein, induces the pre- to post-fusion transition of the S2 subunit, and initiates the membrane fusion.
The prefusion SARS-CoV S1 subunit is structurally organized into 4 separate domains: NTD, CTD1, CTD2 and CTD3 [19]. CTD1 is the receptor binding domain, and one CTD1 in the trimer adopts an up conformation as a prerequisite for the binding of the SARS-CoV to the cellular angiotensin converting enzyme 2 (ACE2) [19]. Similar findings of a protruding up CTD1 have also been documented for MERS-CoV S glyco protein [14, 20]. Through crystal structures of CTD1 in complex with ACE2 has been documented [23], the structure of the trimeric Coronavirus S glyco protein in complex with the cellular receptor has not been documented. The modes involved in the conformational changes of the S glyco protein during Coronavirus infection, mainly for the markedly pathogenic SARS-CoV as well as MERS-CoV, are not totally understood.
Song et al. reported the structure of the SARS-CoV S glyco protein in complex with the host cell receptor ACE2 as shown by cryo-EM [24]. The complicated structure revealed that only one receptor binding domain of the trimeric S glyco protein binds CE2 and takes a protruding up conformation. Additionally, they evaluated the structure of the SARS-CoV S glyco protein and its complexes with ACE2 in separate in vitro conditions that might simulate various conformational states of the S glyco protein at the time of viral entry. Dissociation of the S1- ACE2 complex from certain prefusion spikes was seen and characterized. Further, they characterized the rosette-like structures of the clustered SARS-CoV S2 trimers in the postfusion state seen on electron micrographs. Structural comparisons pointed that the SARS-CoV S glyco protein retains a prefusion architecture following trypsin cleavage into the S1 and S2 subunits along with acidic pH treatment. But binding to the receptor opens up the receptor binding domain of S1, that might aid in the liberation of the S1-ACE2 complex as well as S1 monomers from the prefusion spike and trigger the pre to postfusion conformation transition [24]. The Cryo-EM structure of the 2019-nCoV spike in the pre-fusion conformation is demonstrated in Figure 5 [25].
Recent COVID-19 pandemic around the world is correlated in humans with “severe acute respiratory syndrome” (SARS-CoV2). SARS-CoV2 is an enveloped virus and E proteins present in them are documented to form ion channels, which are basically correlated with pathogenesis. Hence there is an attempt to inhibit these ion channels that in turn might aid in regulating diseases caused by SARS-CoV2 in humans. Based on this, Gupta et al. utilized computational analysis to study the structure along with function of the human “SARS-CoV2E” protein along with its crosstalk with different photochemical [26]. It was found that α-helix as well as loops present in the protein experience random movement under optimal condition, which further manipulates ion channels activity, thus helping the pathogenesis caused through SARS-CoV2 in human as well as other vertebrates. But following binding with Belachinal. Macaflavone E, and Vibsanol E, the random motion of the human “SARS-CoV2E” protein gets decreased, inhibiting the function of the “SARS-CoV2E” protein. It is important to understand that 2 amino acids, i.e. VAL 25 as well as PHE 26, play a crucial role interacting with these 3 phytochemicals. Since these 3 phytochemicals have cleared, the ADMET (Absorption, Distribution, Metabolism, Excretion and Toxicity) property along with Lipinki’s rule of 5’s, they might be used as drugs in controlling disease caused through SARS-CoV2 after more evaluation [26].
DIAGNOSIS
Rapid and proper finding of COVID-19 is important to control outbreaks in the community as well as in hospitals [27]. The present diagnostic tests are reverse transcription-polymerase chain reaction (RT-PCR), real time (rRT-PCR) and reverse transcription loop mediated isothermal amplification (RT-LAMP) [28]. RT-LAMP possesses similar sensitivity to rRT-PCR, is very specific and is utilized to detect MERS-CoV [29]. Laboratory examination includes nasopharyngeal as well as oropharyngeal swab tests. For identification of patients, earlier 2 one step quantitative RT-PCR (qRT-PCR) assays were generated to detect 2 separate regions (ORF1b and N) of the SARS-CoV2 genome [30]. 3 novel RT-PCR assays that targeted the RNA dependent RNA polymerase (RdRp)/Helicase (Hel), spike (S) and nucleocapsid (N) genes of SARS-CoV2 were developed. Of the 3 novel assays, the COVID-19- RdRp/Hel assay had the lowest limit of detection in vitro; highly sensitive and specific assays might aid in improvement of laboratory diagnosis of COVID19-infection [31].The SARS-CoV2 E gene assay had higher sensitivity as compared to RdRp gene assay combined with the one step RT-PCR system [32]. The E gene PCR was enough for diagnosing SARS-CoV2 infection, but the RdRp protocol was recommended for confirmation of a positive result [33]. The total positive rate of RT-PCR detection of SARS-CoV2 infection in 4880 cases from one hospital in Wuhan was 38% [34]. The positive rate of PCR for oropharyngeal swabs is not very high; only 53.3% of COVID-19 confirmed patients had positive oral swab tests [35]. In a series of 51 patients with confirmed COVID-19 infection, 71% patients were RT-PCR positive at the first time of testing of throat swab or sputum samples [36]. The RT-PCR results mostly become positive after several days (2-8) days [37]. Automated solutions for molecular diagnostics can handle huge quantities of samples and may be scaled to keep pace with fluctuating demand [38].
The good analytical performance of a molecular assay for identifying SARS-CoV2 on a high throughput platform, the cobas 6800, was seen with a minimal hand on time, while giving fast and reliable reports [39]. The present laboratory test is time consuming as well as shortage of commercial kits delays diagnosis. For the patients suffering from fever, sore throat, fatigue, coughing or dyspnoea, that is correlated with recent exposure.
COVID19-infection should be diagnosed with typical chest computed tomography (CT) characteristics in spite of negative RT-PCR results [40] (Figures 6, 7). Of 1014 patients, 59% had RT-PCR positive, while 88% had positive chest CT scans [42]. Coronavirus belongs to the Corona viridae family; hence it does not come as a surprise those imaging findings, which are similar to those of SARS-CoV2 as well as MERS-CoV [43]. Typical CT findings are bilateral pulmonary parenchymal ground glass and consolidative pulmonary opacities, sometimes with a rounded morphology and peripheral lung distribution [40]. 86% of patients demonstrated ground glass opacities or consolidation, and > 1 lobe (71%) with bilateral involvement (76%) was present in the 21 initial chest CT scans [44]. Importantly lung cavitations, discrete pulmonary nodules, pleural effusions as well as lymphadenopathy were lacking [44].Lung abnormalities on chest CT scans were more severe about 10 days following the initial onset of symptoms [45].Chest CT scans can be utilized to find the severity of COVID-19. COVID-19 may also present with chest CT scans abnormalities in asymptomatic patients, with rapid evolution from focal unilateral to diffuse bilateral ground glass opacities, which progressed to or coexisted with consolidation within 1-3 weeks. Combining assessment of imaging features with clinical and laboratory findings could promote early diagnosis of COVID-19 pneumonia [46].With the expanding of diagnostic criteria from laboratory testing to chest CT imaging, more than 14000 patients got diagnosed on February 12, 2020.Further for investigating the occurrence of olfactory and gustatory dysfunctions in patients with laboratory confirmed COVID-19 infection.
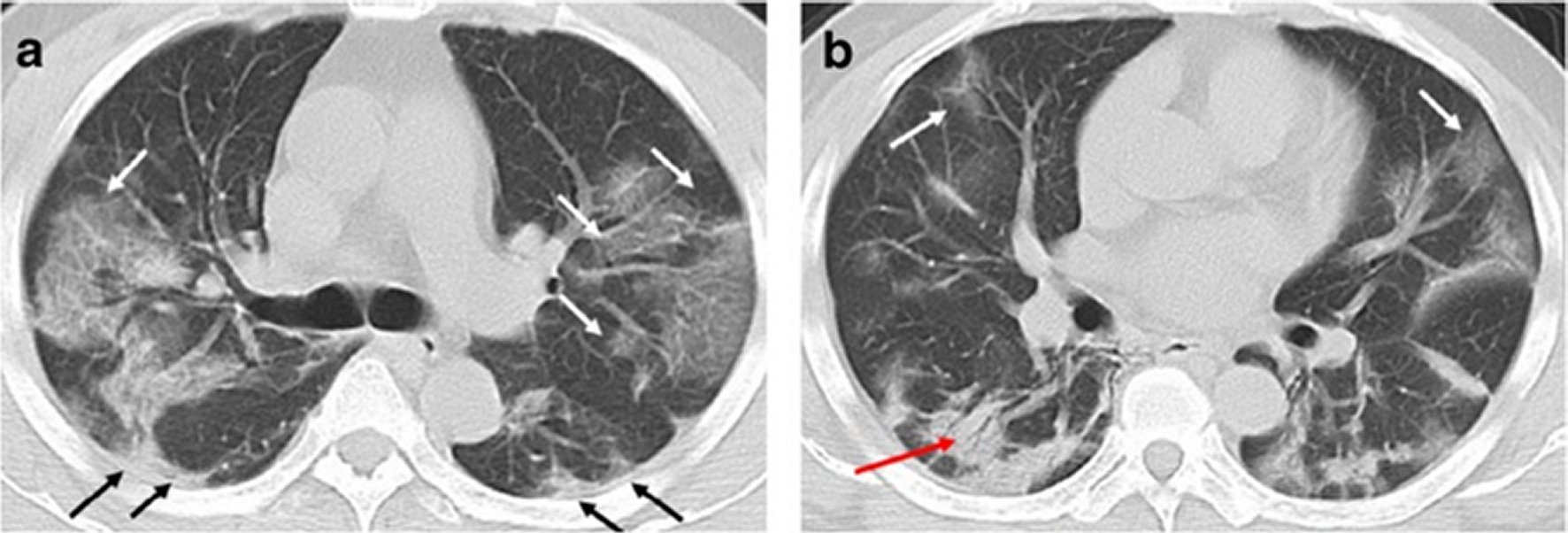
Figure 6. A 49-year-old man with history of recent travel to Wuhan presented with fever and cough for 6 days.a,b: Non-contrast enhanced chest CT showed multiple peripheral patchy ground glass opacities in bilateral multiple lobular and subsegmental with obscure boundary (white arrows), as well as thickening of the adjacent pleura (black arrows). Besides, CT scan also demonstrated consolidation in the right lower lobe and air bronchogram sign in the lesion (red arrow).
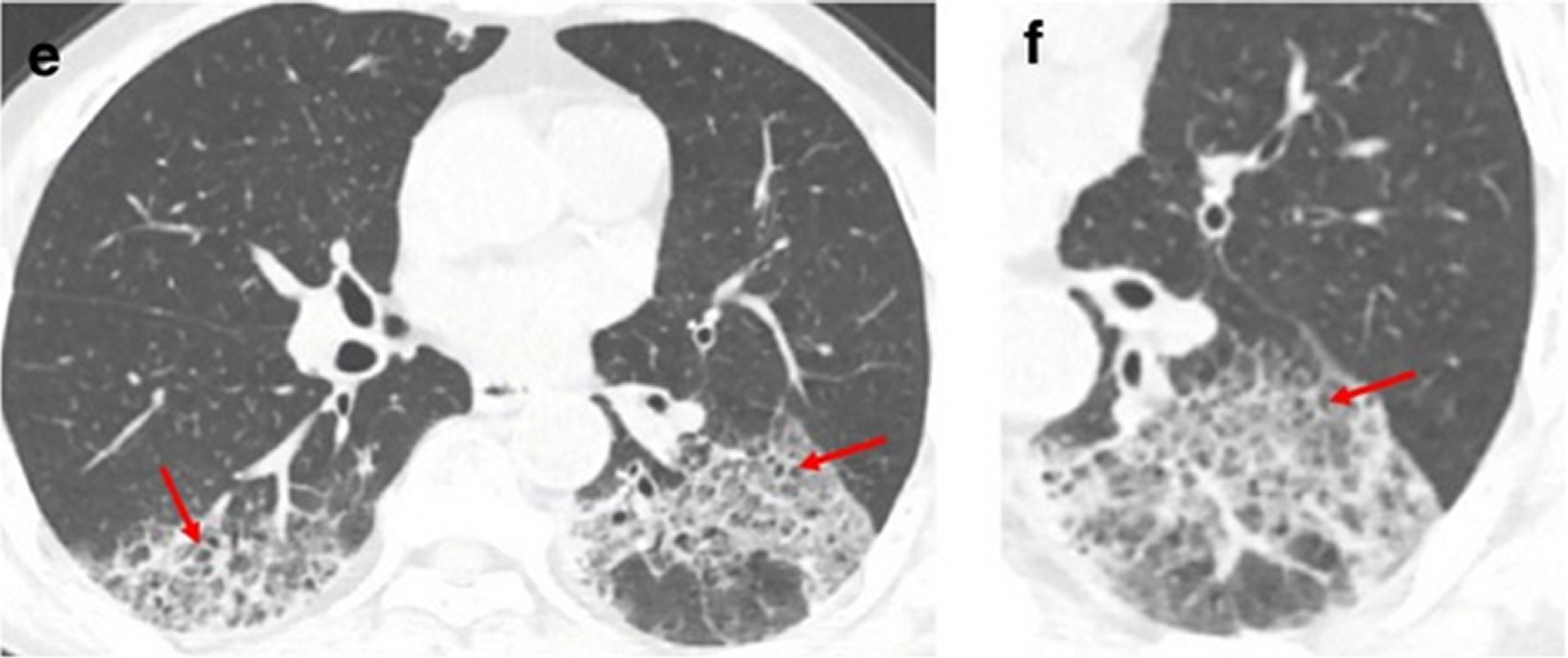
Figure 7. A 62-year-old man with a history of exposure to a market in Guangzhou presented with fever and cough for 11 days.Non-contrast enhanced chest CT showed multiple ground glass opacification in the both lower lobes and thickening of the adjacent pleura (e, f). Red arrows represent the interlobular septal thickening in regions of ground glass opacification (crazy paving pattern).
Lechien et al. recruited patients from 12 European hospitals [47].These epidemiological and clinical outcomes were studied namely age, sex, ethnicity, comorbidities as well as general and otolaryngeal symptoms. Patients completed olfactory and gustatory questionnaire on the basis of smell as well as taste component of the National Health and Nutritional Examination Survey, and the short version of the questionnaire of olfactory disorders-Negative Statements (sQOD-NS). A total of 417 mild to moderate COVID-19 patients completed the study (263 females). The most prevalent general symptoms consisted of cough, myalgia and loss of appetite. Face pain and nasal obstruction were the maximum disease-associated otolaryngological symptoms. 85.6% as well as 88% of patients reported olfactory and gustatory dysfunctions, respectively. There was a significant association between both disorders (p<0.001). Olfactory dysfunction occurred prior to the other symptoms in 11.8% of cases. The sQO-NS scores were significantly lower in patients with anosmia compared to normosmic or hyposmic persons (p=0.001). Of the 18.2% of patients without nasal obstruction or rhinorrhea, 79.7% were hyposmic or anosmic. The early olfactory recovery rate was 44%. Females were significantly more affected by olfactory and gustatory dysfunctions as compared to males (p=0.001). Hence olfactory and gustatory disorders are prevalent symptoms in European COVID-19 patients who might not have nasal symptoms. The sudden anosmia or ageusia need to be recognized by the international scientific community as important symptoms of COVID-19 infection [47].
Further with point of view of treatment a perspective on pathophysiology of COVID-19 in relation to currently available clinical data published in literature needs to be specified. Solaimanzadeh tries to emphasize the problems in COVID-19 related to another respiratory illness, including high altitude pulmonary oedema (HAPE) [48]. Both problems have marked similarities that project pathophysiologic trajectories. Both COVID-19 and HAPE show a reduced ratio of arterial oxygen partial pressure to fractional inspired oxygen with simultaneous hypoxia and tachypnoea. There also seems to be tendencies for low CO2 in both conditions. Radiologic findings of ground glass opacities are present in about 86% of patients with COVID-19 besides patchy infiltrates. Patients with HAPE also demonstrate patchy infiltrates all through the pulmonary fields, mostly in asymmetric pattern, and CT imaging showed escalated lung markings as well as ground glass like changes. Widespread ground glass opacities are mostly a presentation of hydrostatic pulmonary oedema. In the same line, increased fibrinogen levels in both situations are probably an epiphenomenon of oedema generation instead of coagulation activation. Autopsy results of COVID-19 fatality showed bilateral diffuse alveolar damage correlated with pulmonary oedema, proinflammatory concentrates and indications of early-phase acute respiratory distress syndrome (ARDS). HAPE itself is originally caused by an enhancement of pulmonary capillary pressure and stimulates alveolar-capillary permeability through high pulmonary artery hydrostatic pressure, which causes protein rich as well as mildly haemorrhagic oedema. It seems that COVID-19 as well as HAPE both discretely converges on ARDS. Considering this, a countermeasure that has been demonstrated to be efficacious in the analogous problem of HAPE is acetazolamide. Acetazolamide possesses multiple actions on various organ systems, potently decreases hypoxic pulmonary vasoconstriction, and improves minute ventilation as well as expiration vital capacity. Other treatment options to consider are also aimed to reduce pulmonary pressure are nifedipine as well as phosphodiesterase inhibitors [48].Although the virus nucleic acid RT-PCR test has become the standard method for diagnosis of SARS-CoV2 infection, these real time PCR test kits have many limitations [49].Additionally, high false negative rates were reported [49]. An urgent need is there to get an accurate as well as a rapid test method for quickly finding lots of infected patients as well as asymptomatic carriers for avoiding viral transmission. Li et al. have developed a rapid as well as simple point of care lateral flow immunoassay that can find IgG and IgM antibodies simultaneously against SARS-CoV2 virus in human blood within 15 minutes [49]. With this test kit, they conducted clinical studies for validating its clinical effectiveness uses. The clinical sensitivity and specificity for finding were determined utilizing blood samples taken from 397 PCR confirmed COVID-19 cases and 128 negative patients at 8 separate clinical sites. The overall testing sensitivity was 88.86%, while specificity was 90.63%. They examined clinical diagnosis results collected from various types of venous as well as fingerstick blood samples. The results pointed that high detection consistency around samples from fingerstick blood, serum as well as plasma of venous blood. The IgM-IgG combined assay had better utility as well as sensitivity as compared to single IgM or IgG test. It can be used for the rapid screening of SARS-CoV2 carriers, or asymptomatic, in hospitals, clinics as well as test laboratories [49]. Analysis of data from 33 patients with complete clinical course were analysed and levels of blood urea as well as creatinine were higher in nonsurvivors than in survivors [50]. COVID-19 uses the same cell entry receptor like SARS-CoV, ACE2, which regulates both cross-species as well as human-human transmissions [51]. Proximal tubular cells also express higher levels of the ACE2 receptor, which increases susceptibility to COVID-19 [52] and induces renal injury. Analysis of data from 33 patients with complete clinical course was evaluated, and levels of blood urea as well as creatinine were higher in nonsurvivors than in survivors [50]. COVID-19 infection has a clustering onset and has increased chances of affecting older males (average age of 51 years old) with comorbidities [53]. Histological examination of lung biopsy samples revealed bilateral diffuse alveolar damage with cellular fibromyxoid exudates [54]. Proximal tubular cells also express higher levels of the ACE2 receptor, which increases susceptibility to COVID-19 [52] and induces renal injury.
ISOLATION
Usual public health measures, that include isolation, quarantine, social distancing as well as community containment, can be utilized for halting the pandemic by applying lessons that we have learnt from SARS [55, 56]. For the COVID-19 pandemic, China issued the largest quarantine in history. All the residents living in mainland China were locked in as well as in cities. Public transportation, like buses, trains, ferries as well as airports were shutdown. With the trajectory of this outbreak, the Chinese government scaled up lot of efforts for keeping pace with the fast escalation of cases as well as geographical spread. The Wuhan government utilized gyms as well as 2 convention centres, transforming them into makeshift hospitals, having 3,400 beds in one day to isolate COVID-19 patients from healthy controls. More makeshift hospitals were under construction. Isolation beds were rapidly expanded from only 137 at the beginning of the outbreak of COVID-19 to 56,000 to segregate infected patients from no infected persons. This rapid as well as decisive response of China aided in reducing the control reproduction number as well as transmission rate. Due to that, Increase in COVID-19 started slowing down on February 14, 2020. India had an early lockdown just with the detection of a single case. Extensive evaluation of patients coming from other countries was initiated, besides a lockdown of 5 weeks subsequently and rate of spread reduced. In contrast countries like USA, UK and Italy, who did not have an early lockdown, had a massive spread (Table 1). Human Coronaviruses can get effectively inactivated within 1’using surface disinfectant procedures with 62-71% ethanol, 0.5% H2O2 or 0.1% sodium hypochlorite [52].
PREGNANCY WITH COVID-19
The current COVID-19 pneumonia pandemic caused by SARS-CoV2 virus is spreading globally at a rapid rate with a reproduction number (RD) of 2-2.5, pointing that 2-3 persons will be infected from an index patient. Pregnant women with their foetuses represent a high risk population during infectious disease outbreaks [57].Until March 16, 2020, outcome of 55 pregnant women infected with COVID-19 as well as 46 neonates have been documented in the literature, without any definite proof of vertical transmission [57].Physiological as well as mechanical changes in pregnancy enhance susceptibility to infections in general, especially when the cardiorespiratory system is affected, and aid in fast progression to respiratory failure in the gravid. Moreover, the pregnancy bias towards T-helper 2 (Th2) system dominance that protects the fetus, makes the mother having higher vulnerability to viral infections, that are more effectively contained by the Th1 system. These special challenges make it imperative that an integrated approach is done for pregnancies affected by SARS-CoV2 virus [57].
Dashrath et al. presented a review of pathophysiology as well as susceptibility, diagnostic challenge with reverse transcription polymerase chain reaction (RT PCR) assays, therapeutic controversies, intrauterine transmission and maternal-fetal complications. Fetal surveillance due to predisposition to intra uterine growth restriction (IUGR) as well as special attention to labor as well as delivery was addressed. Further, they concentrated on keeping frontline obstetric care providers safe principles of workplace segregation, responsible social distancing, and containment of cross-infection to health care providers, marked use of personal protective equipment judiciously as well as telemedicine. Their objective was to provide a framework that can be used by tertiary maternity units that manage pregnant women in the flux of a pandemic while maintaining the safety of the patient as well as health care provider at its core [57]. No evidence supports adverse birth outcomes, intrauterine infection, or vertical transmission of COVID-19 [58]. However, is known that viral infections can be acquired when the infant passes via the birth canal during vaginal delivery or via postpartum breast feeding [59].
TREATMENT
The SARS-CoV2 virus broke out in December 2019 [60].There have been revisions of the Guidelines the 5th edition of Guidelines advices antivirals like IFN-α, lopinavir/ritonavir as well as ribanivir for treating COVID-19. The particular method for delivering IFN-α is vapouring inhalation at a dose of 5 million units (as well as 2 ml of sterile water for injection) for adults, 2 times/day. Dosage of lopinavir/ritonavir is 400mg/100 mg for adults’ 2 times/day. Ribanivir is given as intravenous infusion in a dose of 500 mg for adults 2-3 times/day combined with IFN-α or lopinavir/ritonavir. Chloroquine phosphate is given orally at a dose of 500 mg (300mg for Chloroquine) for adults 2 times/day. Arbidol is delivered orally at 200 mg dose for adults’. Duration of therapy is not higher than 10 days [60].
IFN-α represents a broad spectrum antiviral used for hepatitis treatment, although it has been documented to inhibit SARS-CoV2 reproduction in vitro [61]. Lopinavir/ritonavir are medications for HIV infection, utilized in combination with other medications for therapy of adults as well as children older than 14 years old who have been infected with HIV [62].Chu et al. observed that lopinavir/ritonavir had anti SARS-CoV2 action in vitro as well as in clinical studies [63]. Ribanivir represents a nucleoside analogue having broad spectrum antiviral actions.111 patients with severe SARS treated with Ribanivir monotherapy as well as 41 patients with SARS treated with lopinavir/ritonavir combined with Ribanivir were compared in a study [64].They observed those receiving combined therapy had lower risk of ARDS as well as death [64]. Chloroquine is used widely as an ant malarial and was detected to have potent broad-spectrum antiviral activities in 2006 [65]. Chloroquine was observed to block SARS-CoV2 infection at low micromolar levels, with a half-maximal effective value (EC50) of 1.13 µM and a half cytotoxic value (CC50) higher than 100 µM [66]. Arbidol represents an antiviral which can efficiently inhibit SARS-CoV2 infection at a level of 10-30 µM in vitro [67].
Besides these drugs used in above Guidelines, favipiravir is a drug that needs more lime light. It got approved for the therapy of novel influenza on February 15th, 2020, in China. This drug is undergoing clinical trials for treating COVID-19. Favipiravir is a new kind of RNA-dependent RNA polymerase (RdRp) inhibitor. Besides its anti influenza action, favipiravir can block the replication of flavi, alpha, filo, bunya, arena, noro as well as other RNA viruses [68]. Favipiravir gets converted into an active phosphoribosylated form (favipiravir-RTP) in cells and gets detected as a substrate by viral RNA polymerase, thereby inhibiting RNA polymerase activity [69]. Hence favipiravir might have potential anti viral activities against SARS-CoV2. A clinical trial on favipiravir for the therapy of COVID-19 was started by the Clinical Medical Research Centre of the National Infectious Diseases, started on February 14, demonstrated promising outcomes. Initial results via a total of 80 patients (both experimental as well as control group) pointed that favipiravir had increased potent viral activities as compared to lopinavir/ritonavir [70]. No significant side effects were observed in the favipiravir treated group, having significantly less side effects as compared to lopinavir/ritonavir group [70].
Remdesivir is another drug which has potential benefits for treating COVID-19. It is a nucleoside analogue as well as broad spectrum antiviral. Animal experiments [71] pointed that Remdesivir can efficaciously reduce the viral load in lung tissue of mice infected with MERS-CoV, improve lung function as well as ameliorate pathological damage to lung tissue. Remdesivir was observed to potently blocks SARS-CoV2 infection at low micromolar levels having an increased selectivity index (half maximal effective concentration (EC50) of 0.77 µM; and a half cytotoxic value (CC50) higher than 100 µM; SI >129.87) [66]. In USA, outcomes documented of Remdesivir by Holshue et al. were promising in treating a patient of COVID-19 [72]. For analysis of efficiency as well as safety of this drug, a randomized, placebo controlled, double-blind, multicenter phase III clinical trial was started on February 5, 2020, in Chin [73, 74]. Experimental group patients got an early dose of 100 mg during 9 consecutive days through intravenous infusion, besides the routine therapy. Control group patients got routine therapy as well as same dose of a placebo. The trial is expected to finish by end of April 2020. Currently, Remdesivir have been used maximum in treating severe patients with respiratory involvement needing ventilator [74].
Further studies have also demonstrated that darunavir, which is a second generation of HIV1 protease inhibitor, inhibited SARS-CoV2 infection in vitro [59]. Cell experiments pointed that darunavir inhibited viral replication markedly at a level of 300 µM in vitro, being its inhibition efficiency 280 times more effective as compared to well control group [59]. Other potential drugs were type II transmembrane serine proteases (TMSPSS2) inhibitors as well as BCR-ABL kinase inhibitor, such as imatinib. Hoffmann et al. suggested that SARS-CoV2 utilizes SARS-CoV2 receptor ACE2 as well as cellular proteases TMSPSS2 for entering the target cells. A TMSPSS2 inhibitor should block the entry and hence be a treatment method [75]. Imatinib has anticoronal activity mainly as it inhibits the fusion of virions with the endosomal membranes [76].
A research team of the Shanghai Institute of Material Medica did drug screening in silicon as well as enzyme activity test, revealing 30 compounds which have potential anti viral activities against SARS-CoV2 [20]. These compounds were indinavir, saquinavir, lopinavir, carfilzomib, ritonavir, atazanavir, darunavir, tipranavir, fosamprenavir, enzaplatovir, presatovir, abacavir, bortezomi belvitegavir, maribavir, raltegavir, montelukast, deoxyrhapontin, polydatin, chalcone, disulfiram, carmofur, shikonin, ebselen, tideglusib, PX-12, TDZD-8, cyclosporine, and cinanserin. They also observed that Chinese herbal medicines might contain active ingredients against SARS-CoV2 [77].
IVERMECTIN
Ivermectin is an FDA-approved broad spectrum antiparasitic agent [78], which recently has been shown to have antiviral activity against a wide range of viruses in vitro [79]. Initially, was found as an inhibitor of interaction between the HIV-1 integrase protein (IN) and the importin (IMP) α/β1 heterodimer that causes IN nuclear import [80]. Since then, Ivermectin has been confirmed to inhibit IN nuclear import as well as HIV-1 replication [81]. Although other actions of Ivermectin have been documented, Ivermectin has been demonstrated to inhibit nuclear import of host-like [82] as well as viral proteins, including simian virus SV40 large tumor antigen (T-ag) as well as dengue virus (DENV) non-structural proteins 5 [80]. Significantly, it has been shown to halt infection by RNA viruses like DENV1-4 WNV [83], Venezuelan equine encephalitis virus (VEEV) as well as influenza [84]. This broad spectrum activity has been thought to be secondary to the reliance by many separate RNA viruses on IMP α/β1 at the time of infection [85]. Similarly Ivermectin has been revealed to be efficacious against the DNA virus psudorabies virus (PRV) both in vitro as well as in vivo, with Ivermectin therapy increasing survival in PRV infected mice [86]. This efficacy was not seen for Ivermectin against Zika virus (ZIKV) in mice, although the authors said that study limitations warrant re-examination of Ivermectin’s anti-ZIKV activity [87]. Further, Ivermectin was the focus of a phase III clinical trial in Thailand in 2014-2017, against DENV infection, in which a single daily oral dose was observed to be safe and caused a marked decrease in serum values of viral NS1 protein, but no difference in viremia or clinical advantage [88].
The aetiological agent for the recent COVID-19 pandemic, SARS-CoV2, is a single stranded positive sense RNA virus, which is closely related to ’severe acute respiratory syndrome’ coronavirus(SARS-CoV). Studies on SARS-CoV proteins have shown a potential role of IMP α/β1 at the time of infection in signal-dependent nucleocytoplasmic shutting of the SARS-CoV nucleocapsid protein [89], which might influence host cell division [90]. Additionally, the SARS-CoV accessory protein ORF6 has been revealed to antagonize the antiviral activity of the STAT1 transcription factor by sequencing IMP α/β1 on the rough ER/Golgi membrane [91]. These reports pointed that Ivermectin’s nuclear transport inhibitory activity might be efficacious against SARS-CoV2.
For testing the antiviral activity of Ivermectin towards SARS-CoV2, Leon Caly et al. [92] infected Vero/hSLAM cells with SARS-CoV2 isolate Australia/VICO1 at an MOI of 0.1 for 2 h, followed by addition of 5 µM of Ivermectin. Supernatant and cells pellets were harvested at days 0-3 and examined by RT-PCR for the replication of SARS-CoV2 RNA. At 24 h, there was a 93% decrease in viral RNA present in supernatant (pointing to released virions) of samples treated with Ivermectin as compared to vehicle DMSO. On the same hand, a 99.8% decrease in cell-correlated with Ivermectin therapy. After 48 h, this action escalated to an ~5000 times decrease of viral RNA in Ivermectin treated samples as compared to control samples, pointing that Ivermectin therapy caused an efficient loss of essentially all viral material. Agreeing with this idea, no future decrease in viral RNA was present in 72 h. As previously observed [69], no toxicity of Ivermectin was detected at any of the time points tested, in either the sample wells or in parallel tested drug alone samples. To further find the efficacy of Ivermectin, cells infected with SARS-CoV2 got treated with serial dilutions of Ivermectin 2 h post infection as well as supernatant and cell pellets collected for real time RT-PCR at 48 h. Just like above, a higher than 5000 times decrease in viral RNA was observed in both supernatant and cell pellets from samples treated with 5 µM Ivermectin at 48 h, equating to 99.98% decrease in viral RNA. No toxicity was observed again with Ivermectin at any concentratation evaluated. The IC50 of Ivermectin therapy was found to be ~2 µM. Emphasizing the fact that the assay actually found SARS-CoV2, RT-PCR experiments were repeated with primers specific for the viral Rd Rp gene rather than the E gene (above) with nearly similar results seen for both released (supernatant) as well as cell associated virus. Taken together these results, it is suggested that Ivermectin has antiviral activities against SARS-CoV2 clinical isolates in vitro; with a single dose able to control viral replication within 24-48 h in their system [92].More work is required with higher doses and safety in pregnancy.
VACCINES
The structure of SARS-CoV2 S protein has been attained, which should help in the rapid generation as well as examination of medical countermeasures for tackling the continuing public health crises [24]. Most of the vaccines getting developed for Coronaviruses target the spike glycoproteins or S proteins [93].
Vaccines formation is a long process, and with no vaccine available at the time a pandemic outbreak. Like in the Ebola endemic that took place in 2013, Ebola vaccine was selected for phase 1 clinical trial for its safety as well as immunogenicity in Africa as well as Europe [94]. In November 19, the European Union granted marketing authorization to Merck Sharp as well as Dohme BV for their Ebola Vaccine, Ervebo. Luckily Moderna Company announced on February 24, 2020, that the company’s experimental mRNA COVID-19 vaccine, known as mRNA-1273, is ready for human testing. It is considerably fast development cycle to generate an initial vaccine just weeks following identification of the SARS-CoV2 genetic sequence. The clinical trial of safety as well as immunogenicity of mRNA-1273 in the therapy of COVID-19 is under investigation (Clinical Trials.gov Identifier NCT 04283461). Further, a new oral SARS-CoV2 vaccine has been successfully generated at Tianjin University, which utilizes food grade safe Saccharomyces cerevisiae as a carrier and targets the S protein. There are 18 biotechnology companies as well as universities in China working on SARS-CoV2 vaccine. Vaccines for SARS-CoV2 have been formed much faster as compared to those of Ebola vaccine due to the collaborative work of scientists around the world as well as fast track approval of SARS-CoV2 Vaccine development efforts by the Chinese government. Even with Indo US collaboration efforts are being made to develop vaccines, some in trials till animal level, while development of nasal vaccines are also ongoing. Zhang et al detailed on the progress as well as our prospects on the field of vaccine developed against SARS-CoV2 [95].
CONVALESCENT PLASMA TRANSFUSION
Convalescent plasma transfusion was delivered early following symptoms onset in the therapy of SARS and the pooled odds of mortality after treatment (odds ratio 0.25) [96]. However in Ebola virus disease, the transfusion of 500 ml of convalescent plasma in 84 patients was not correlated with a marked improvement in survival [97]. In a laboratory test, the COVID-19 virus was detected from the bronchoalveolar lavage fluid of a critically ill patient, and it could get neutralized by sera from various patients [98]. The National Health of China appealed to convalescent patients to donate blood for the therapy of COVID-19 infection. Convalescent plasma needs to be collected within 2 weeks following recovery time to make sure a high neutralization antibody titer is recovered. The problem in getting plasma in the convalescence limits its clinical application. Well designed clinical trials are required to further examine the effectiveness as well as safety of convalescent plasma transfusion in patients with COVID-19 infection.
CONCLUSION
While the origin of COVID-19 is still being investigated, COVID-19 has features typical of Coronaviriridae family, and was classified in the beta Coronavirus 2b linage. COVID-19 can be transmitted between humans. Interventions that include intensive contact tracing followed by quarantine as well as isolation can efficiently decrease the spread of COVID-19, with the effect of travel restrictions. Wear masks, washing hands as well as disinfecting surfaces aid in decreasing the infection. Human Coronaviruses can get effectively inactivated within 1’using surface disinfectant procedures with 62-71% ethanol, 0.5% H2O2 or 0.1% sodium hypochlorite.
Diagnosis using RT-PCR or antibodies is very important to decide the appropriate therapy. Typical CT scan findings can help early screening of suspected cases and diagnosis of COVID-19. COVID-19 infection has a clustering onset and has increased chances of affecting older males (average age 51 years old) with comorbidities
The commonest symptoms were fever, cough, expectoration, headache, myalgia or fatigue, diarrhea and haemoptysis. Some people might experience SARS. Other organs are also susceptible to COVID-19. The single cell RNA-seq data was utilized for evaluating ACE2 expression for telling the potential risk of various human organs to COVID-19 infection uses the same cell entry receptor like SARS-CoV, ACE2, that regulates both cross-species as well as human-human transmissions. Analysis of data from 33 patients with complete clinical course revealed that levels of blood urea as well as creatinine were higher in nonsurvivors than in survivors. The pandemic is actively continuing. Different novel treatments are getting tried including remdesivir, favipiravir, hydroxychloroquin with azithromycin, ivermectin and in serious case acetazolamide, nifedipine along with PDE Inhibitors. Further convalescent plasma is being tried in USA with 3 successful outcomes till date on 12/4/2020 along with other countries. Efforts are being made to get a vaccine as early as possible and lockdown the infection. The social distancing contact tracing and early testing to reduce the spread and mortality till the whole world can come to terms in getting over this pandemic by containing the virus besides use of extra sanitation measures use of masks gloves, PPE are now the main keys to prevent the infection. God willing we will soon be able to conquer this virus with the collaborative worldwide efforts of doctors, scientists, governments, and paramedical personnel.