1. INTRODUCTION
Multiple drug resistant (MDR) bacteria present an enormous challenge for the medical community, in which bacteria develop resistance against more than one antibiotic drug agent. The World Economic Forum Global Risks present data uncovers the greatest growing danger to our health, currently, is antibiotic resistance [1]. The World Health Organization (WHO) and the Centers for Disease Control (CDC) have communicated their immense concerns for the rising rate of antibiotic resistance [2]. In the 2019 Centers of Disease Control and Prevention Antibiotic Resistant Threats Report, it was confirmed that antimicrobial resistant pathogens caused 2.8 million infections with more than 35,000 deaths each year in the United states between the years of 2012 and 2017 [3]. Infections due to antibiotic resistance cause approximately 600 deaths annually from the exposure to bacterial pathogens in many healthcare centers such as hospitals [4]. However, multiple drug resistant (MDR) Gram-negative bacteria cause a majority of bacterial infections. The Centers for Disease Control have confirmed that Acinetobacter baumanii, a Gram-negative bacterium, causes a majority of hospital infections [5]. A. baumanii resides in hospitals and are highly contagious pathogenic bacteria.
Gram-negative bacteria have a higher rate of antibiotic resistance because of the Gram-negative bacterial outer membrane that acts as a natural physiological barrier of resistance against antibiotics. Gram-negative bacteria have a cell envelope that consists of an outer membrane, a periplasmic space, and an inner membrane. The three layers of the cell envelope allows Gram-negative bacteria to resist a broader range of antimicrobial drug agents and compounds. For instance, any changes in the outer membrane of Gram-negative bacteria, such as altering its hydrophobic components or mutations of genes required for the protein expression of porins, can develop into resistance [6]. Gram-negative bacteria also have many additional mechanisms of resistance, such as DNA mutations of antimicrobial targets; degrading antimicrobials and modifying antimicrobials; acquiring genetic components such as plasmids, transposons or integrons; altering the components of the cell wall and modifying lipopolysaccharides; decreasing the expression of porins, which reduces the uptake of antimicrobials; and the overexpression of efflux pumps [2]. Therefore, selecting effective antibiotics to treat bacterial infections, generated by resistant and pathogenic bacteria, has been immensely challenging [3].
Treating MDR bacterial infections is arduously challenging because many MDR bacteria overexpress efflux pumps [2]. Overexpression of multiple drug resistant efflux pumps or MDR efflux pumps allows the massive export of a broad range of antibiotic compounds [2]. Gram-negative bacteria utilize all the classified families of efflux pumps, such as the ABC superfamily; the major facilitator superfamily (MFS); the multidrop and TolC compound extrusion (MATE) family; the small multidrug resistance pumps (SMR) family and the Resistance Nodulation Division (RND) family [2]. However, Gram-negative bacteria mainly use RND efflux pumps for MDR efflux activity [Anes]. RND pumps are accountable for the expulsion of a broad range of agents and compounds. RND pumps include homotrimers termed as the AcrAB-TolC efflux pump [7]. The AcrAB-TolC subunits are transmembrane proteins that are oriented into the inner membrane, outer membrane, and through the periplasmic space. Its structure is a triplex with protein subunits termed AcrA located in the periplasmic space, the TolC is oriented in the bacterial outer membrane, and AcrB resides in the inner membrane region of a bacterial cell [7] (Figure 1). The binding sites of AcrAB-TolC, a homotrimer efflux pump, can bind to and attach to many antibiotics with different sizes and chemical properties [8]. MDR efflux pumps can expel a wide range of antibiotics, which contributes to the continued proliferation of MDR Gram-negative bacteria.

Figure 1: The AcrAB-TolC Efflux Pump, RND antibiotic efflux pump, ex-ports antibiotic drugs from the cytoplasmic space of a bacterial cell, which traverses through the inner membrane, into the periplasm, and then exits the bacterium’s outer membrane via the AcrB, AcrA, and TolC subunits of AcrAB-TolC efflux pump, respectively.AcrB is located in the inner membrane and periplasm while AcrA is implanted in the periplasm. TolC is oriented into the periplasm and outer membrane. The presence of antibiotics triggers the AcrAB complex to alter its conformation in order to retrieve and recruit the TolC subunit. The opening of TolC remains closed until the antibiotic expulsion in order to isolate the periplasm from the external environment. Lastly, the fully assembled AcrAB-TolC pump contracts and opens to expel the antibiotic drug through its chamber. The AcrAB-TolC then closes following the expulsion of the antibiotic drug molecule.
In contrast, manipulating the metabolism of bacteria can increase the uptake of antibiotics. Metabolites in high concentrations increase bacterial antibiotic resistance. Metabolites such as indole, ammonia, nitric oxide, and hydrogen sulfide can amplify antibiotic resistance [9]. However, carbon in higher amounts eradicates more pathogenic bacteria by increasing antibiotic sensitivity. The high influx of carbon from the environment surrounding bacteria controls antibiotic sensitivity by raising nicotinamide adenine dinucleotide (NAD) + hydrogen (H) or NADH activity and increasing the proton motive force (PMF) [10]. The rise of NADH levels and the rapid activity of PMF can both activate the TCA cycle of bacterial carbon-dependent metabolism [10]. Upregulating the expression of proteins as a part of bacterial metabolic pathways can amplify the rate of the PMF, which can intensify antibiotic sensitivity. PMF also powers the efflux activity of many RND pumps. By inhibiting or deleting the genes that code for the expression of efflux pump proteins, the PMF can decrease, thereby disrupting efflux activity.
Although many novel antibiotics have been developed to treat resistant bacterial infections, many pathogenic bacterial strains persistently resist many of these novel agents [3]. Designing novel and alternative antibiotics that are gene-based could combine novel genetic engineering techniques with antibiotic development to combat antibiotic resistance. However, to overcome the antibiotic resistance caused by drug efflux, the current literature available needs a more succinct elucidation of the fundamentals of drug efflux mechanisms [2]. An understanding of drug efflux mechanisms, such as the regulation and response to antimicrobials, is urgently required for combating antibiotic resistance [2]. For these reasons, this review attempts to accurately describe the drug efflux mechanisms of the RND AcrAB-TolC efflux pump through a genomic and metabolomic perspective of antibiotic efflux and influx. This review and perspective will discuss the reduction of antibiotic resistance by inhibiting drug efflux protein pump expression and stimulating bacterial metabolic activity to increase antibiotic influx. The overall purpose of this review is to present an alternative gene-based antimicrobial strategy against resistant bacteria that may increase the antibiotic retention in bacterial cells by inhibiting antibiotic efflux and metabolically amplifying antibiotic influx.
2. BACTERIAL ANTIBIOTIC RESISTANCE
Bacteria can adapt well too many different environmental conditions partially because of its ability to select advantageous DNA mutations that allows bacterial cells to survive, reproduce, and dominate its population. Antibiotic resistance genes can be transferred between bacterial cells and limit the bacterial susceptibility of antibiotics. Bacteria become multidrug resistant through the accumulation of antibiotic resistance genes that are transferred by plasmids or transposons [11]. Each resistant gene may code for the inhibition of a drug agent or encode multidrug efflux pump activity [11]. Bacteria develop antibiotic resistance by the transference of antibiotic resistance genes through a horizontal gene transfer [12]. Bacteria have three modes of horizontal gene transfer such as conjugation (bacterial sexual reproduction), transformation (incorporating naked DNA), and transduction (mediated by phages) [13]. Bacterial cells receive antibiotic resistant genes and after gene expression the diffusion of antibiotics through bacterial cell pores is prevented, causing antibiotic efflux; the antibiotics’ binding to antimicrobial target sites is blocked; the antibiotics are targeted for hydrolysis and inactivation; and changes in metabolic pathways can occur and alter regulatory networks [13].
The addition of antibiotics to a bacterium’s environment forces the bacterium to select gene mutations that give it an advantage. Bacteria can acquire these DNA mutations, which allows the bacterial cells to continue reproducing in the presence of antibiotic drugs. Bacteria carrying advantageous DNA mutations of antibiotic resistance can reproduce without much competition from the dead bacteria that lacked the advantageous mutations. After the exposure to bactericide antibiotics, the population of bacteria that survived are called persister cells. The persister cells not only survive but also are key determinants for further initiation of antibiotic resistance [14]. Consequently, natural selection is the central component and the driving force for the development of antibiotic resistance in bacteria [15] where exposure to antibiotics impels the selection of antibiotic resistance genes that then become rapidly disseminated between many bacterial cells.
Pathogenic bacteria can become resistant to antibiotics through acquiring antibiotic resistant genes that code for degrading antibiotics, using efflux pumps, and shielding targets from binding to antibiotics. For example, bacteria can acquire DNA from its environment and transfer antibiotic resistant genes called ‘mosaic’ genes [16]. Streptococcus pneumoniae develops antibiotic resistance to penicillin by sharing mosaic penicillin-binding protein genes or pbp genes between bacteria [17]. The mutations in pbp genes alter proteins such as enzymes to exhibit less affinity for penicillin. The mosaic pbp genes originated from the continued mixture and rearrangements of DNA by the bacteria named Streptococcus mitis [18]. The influx of a class of antibiotics known as cefiximes, which are third-generation cephalosporins, has also been blocked after mosaicism and point mutations in penA genes that code for Pbp expression in N. gonorrhoeae [19]. Additionally, A. baumanii becomes antibiotic resistant by obtaining resistant genes from conjugation, which is mostly a horizontal transfer of genes, and via mutations in its DNA. The enzymes of acetyltransferase and the nucleotidyltransferases modify the antibiotics called aminoglycosides that form resistant A. baumannii bacterial strains [5]. A. baumanii inserts mutations into the genes gyrA and parC. Then, enzymes become overexpressed to alter the antibiotic target called 16S rRNA and prevents the aminoglycoside to bind to 16S rRNA, inhibiting 16S rRNA activity. Another example of antibiotic tolerance includes propionate, a volatile fatty acid (VFA), which may become toxic for Escherichia coli cells, yet E. coli can develop tolerance of propionate by utilizing propionate as a carbon source for generating energy. The volatile fatty acid or VFA called propionate can inhibit many metabolic processes when the VFAs amass in high concentrations within a bacterial cell and can assist with eliminating antibiotic resistant bacteria [20]. However, E. coli cells modify the mRNA transcripts of genes needed for metabolizing propionate, and then the anaerobic metabolism of propionate is amplified.
The bacterial antibiotic resistance of quinolone is an example of antibiotic resistance caused by the destruction of antibiotics and through blocking the binding of antibiotics to bacterial targets. Quinolone can effectively block the expulsion of antibiotic drugs from bacterial cells, and copies of quinolone antibiotics have been derived to inhibit the efflux pump called NorA in S. aureus [21, 22, 23, 24, 25, 26, 27, 28, 29, 30]. However, the resistance to quinolone antibiotic drugs is propagated through the gene expression of qnr. The qnr mRNA transcript is translated into QnR proteins that consist of pentapeptide repeat proteins or PRPs [10]. The PRPs binding to DNA gyrase and to topoisomerase IV restricts the quinolones inhibition of DNA gyrase and topoisomerase IV. When the quinolone antibiotic attempts to attach, the PRPs begin to bind to the DNA and inhibits the quinolones’ binding to the enzyme-to-DNA interfaces of topoisomerase IV and of the DNA gyrase [31]. The PRPs prevent the quinolones from converting DNA gyrase and the topoisomerase IV into toxic enzymes that degrade bacterial chromosomes, allowing the topoisomerases to continue completing the activities of rebuilding dsDNA breaches, eliminating knots in chromosomes, and relaxing torsional stress [31]. Furthermore, many bacterial enzymes can also hydrolyze and destroy antibiotics, such as Beta-lactams, aminoglycosides, phenicols, and macrolides. Therefore, the continued overuse of antibiotics will only increase the proliferation of resistant bacterial strains [32]. For these reasons, elucidating the functions that establish and proliferate antibiotic resistant bacteria is required for advancing the pharmaceutical design of novel antibiotics that can accurately target antibiotic resistant genes and the cellular properties of drug resistant bacteria.
3. COMBATING BACTERIAL ANTIBIOTIC RESISTANCE
Additional alternative strategies that can effectively combat antibiotic resistance are still presently and urgently needed. A possible alternative method to combat antibiotic resistance was developed by Ayhan et al. that constructed antisense RNA (asRNA) molecules attached to the mRNA transcripts of antibiotic resistant genes. Ayhan et al. designed phosphorodiamidate morpholino rings connected by a phosphorodiamidate (PMO) backbone [33]. Each ring of the PMO was bound to an antisense nucleotide base and blocked the translation of antibiotic resistant genes via steric hindrance [33]. The site of steric hindrance was tightly adjacent to the ribosome binding sequence of the bacteria that obstructed the entrance of bacterial mRNA into ribosomes. Adding peptides to the Peptide-Conjugated Phosphorodiamidate Morpholino Oligomer or PPMOs enhanced its entrance into bacterial cells during conjugation [33]. The phosphorodiamidate, a PMO, backbone impedes the nuclease activity that would disassemble the PPMO. PPMOs present a few issues, which need more research in the area of pharmacokinetics for effectively combining PPMOs with antibiotics [33]. There are issues of mismatched base pairing, determining the position and size of the oligomer, and the effectiveness of PPMOs requires further examination.
Additionally, targeting quorum sense signalling between bacterial cells could provide another highly effective method for reducing the antibiotic resistance of Salmonella typhimurium. When comparing mutant Salmonella typhimurium versus its wild type, the wild-type showed activation of secondary molecules that continued the expression of antibiotic resistant genes under stress and increased pressure. However, the mutant form was not as regulated and did not activate secondary molecules. S. Typhimurim passes through the M cells of the epithelial lining of the intestines, leading to a more systemic infection. The systemic infection caused by S. Typhimurium depends on the virulent genomic region called the S. Typhimurium pathogenicity island or SPI-1 and SPI-4. The SPI-1 and SPI-4 are less regulated in its mutant form, which lessens the frequency of bacteria crossing the M cells of the gastrointestinal tract (GIT) [34]. SPI-2 acts to fortify bacterial cells to evade phagocytosis by macrophages. A mutated gene that encodes the PhoPQ protein decreases expression of the LsrACDB protein complex, which then decreases quorum sensing (QS). When upregulating the gene expression of SPI-1, the gene expression of lsrACDB gradually increases [34]. The quorum sense signaling of a S. Typhimurium infection is completely eradicated after mutation of the gene that encodes the PhoPQ protein. A possible strategy could include an attenuated S. Typhimurium can become re-engineered by adding a QS detecting promoter site with a small interfering RNA (siRNA) or asRNA sequences of lsrACDB to a plasmid vector. The plasmid vector would consist of a pluxR promoter site to detect QS molecules and then release the small interfering RNAs (siRNAs) or asRNA for blocking the transcription of lsrACDB where LsrACDB expression can be reduced, interrupting the quorum sense signalling between S. Typhimurium bacterial cells.
The phytonutrients of plants can potentially reduce antibiotic resistance. Possible alternatives to reduce antibiotic resistance may include the use of plants that release cytotoxic phytonutrients and provide immunity against foreign bacterial cells. For example, reserpine is a plant alkaloid extracted from the roots of the petitiva plant. Reserpine inhibits the efflux pump of Gram-positive bacteria known as Bacillus subtilis. Reserpine binds to the pumps that express three amino acids with the side chain residue groups of phenylalanine and valine. Reserpine causes a 4-fold decrease in the antibiotic resistance of methicillin-resistant Staphylococcus aureus or MRSA caused by S. aureus. Berberine has a MIC of 256 mg/L, but when the NorA pump is active, the berberine MIC is reduced to 16 mg/L [4]. Other examples of plant phytonutrients include polyphenol blockers found in green tea extracts that can inhibit MRSA resistance. Adding 20 mg/L of polyphenols to the antibiotics norfloxacin resulted in a 4-fold decrease in MIC [4]. However, there are not many studies that focus on the medicinal properties and benefits of plant alkaloids [4].
Furthermore, novel synthetic antibiotics have also been developed to decrease antibiotic resistance. Novel antibiotics such as capistruin and malleilactone block bacterial cell transformation. Capistruin blocks bacterial RNA polymerase activity. Malleilactone interacts with quorum sensing molecules. However, Staphylococcus aureus has a lysine at position 164, resulting in antibiotic resistance of thailandamide [35]. The antibiotic of bactobulin can connect to the LuxI-LuxR proteins responsible for quorum sensing [35, 36, 37, 38]. The LuxI-R linkage to bactobulin stimulates a molecule that inhibits bacterial translation in the ribosome. In addition, a list of criteria and properties are needed for establishing a database to help identify and examine the environment for highly pathogenic resistant bacterial types [37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48]. Additional procedures that avoid and prevent bacterial antibiotic resistance include the following: regulate the antibiotics given to animals, perform future research that elucidates the mechanisms of antibiotic resistance and the monitoring of resistance [49], improve the laws and regulations to reduce the prevalence of antibiotic resistance, amplify the research of more novel therapies [50], and provide more aid to developing and low-income countries to more effectively regulate issues of antibiotic resistance [50]. Currently, WHO has an antimicrobial surveillance system that collects data to help reduce the rate of antibiotic resistance [CHokshi]. Reducing the use of plastic items may also block the interaction between microplastics and antibiotic resistance genes in aquatic environments [51]. However, Hall et al. concluded that antibiotic resistance originates from a genetic source [52]; therefore, the solution for eliminating multidrug resistant bacteria may require a gene-based approach. Modifying the genes that encode the binding sites of MDR efflux pumps such as the RND AcrAB-TolC efflux pumps can limit efflux activity and reduce antibiotic resistance.
4. RND-EFFLUX PUMPS AND ANTIBIOTC-RESISTANCE
Many genes coding for the protein synthesis of RND pumps can be shared between bacterial cells through conjugation [53]. The mutations of genes encoding for increased efflux activity are found in the regulatory portion of DNA sequences that modulate the translation and synthesis of efflux pump complexes. The expression of efflux pumps on the bacterial cell surface is altered by the increased presence of gene mutations in an adjacent promoter site. Many DNA mutations in an adjacent promoter site can affect the production of transcription factors that control the cell surface expression of efflux pumps. For example, a point mutation approximately 10 base pairs upstream of the mtrC gene of Neisseria gonorrhoeae increases the amount of activity at the promoter, resulting in amplified expression of efflux pumps to rapidly remove antibiotic drugs [54]. Genes encoding the expression of MDR efflux pumps can also become incorporated into plasmids and then transferred among bacteria. For example, Resistance Nodulation Division (RND) pumps are transported via IncH1 plasmids isolated from the Citrobacter freundii bacteria, which transmits the New Delhi metallo-Beta-lactamase 1 or NDM1-34 for enzymatic decay of antibiotic drugs [55].
As a result, MDR efflux is responsible for amplified antibiotic resistance, which exports antibiotics from bacteria. The antibiotics are pumped and exported from the cytoplasm of the bacteria. The AcrAB-TolC efflux pumps are a part of the RND family of efflux pumps and consist of three protein subunits. The three subunits include AcrA, the mid-transmembrane part of the pump, AcrB located in the bacterial inner membrane, and TolC is positioned in the region of the outer membrane (Figure 1). The AcrAB-TolC pump is assembled in a sequential order [7]. The first step of the AcrAB-TolC assembly includes the interaction between AcrB and AcrA, which then forms the AcrAB bipartite complex [7]. Secondly, the conformation of AcrA changes to then engage and recruit TolC [7]. When the TolC becomes bound to the AcrAB complex, the completely assembled AcrAB-TolC pump enters and remains in a resting state. Thirdly, when AcrB engages a drug, the fully assembled pump opens its conformation and then a constraction expels the drug from the channel into the external environment of the cell [7]. Through observation of tomography results and data collected in vivo, it was found that AcrB and AcrA interact to form into a bipartite complex [7]. The AcrAB bipartite complex stabilizes the AcrAB-TolC efflux pump’s orientation into the bacterial cell envelope [7].
The membrane fusion protein of AcrA connects the pump of the inner membrane to the pump located in the external lipid membrane region where the TolC channel resides. The N-terminus of AcrA anchors itself into the inner membrane through its α-hairpin interaction with the peptidoglycan layer [7]. The hairpin of AcrA is helical and forms an alpha-helical barrel during the shift from no ligand-bound to a ligand-bound state during the assembly of AcrAB-TolC [7] (Figure 1). The AcrAB-TolC protein channel pump does not require substrates with a specific molecular weight, net charge, or surface area. However, mutations in the AcrA gene can weaken the antibiotic resistance of bacteria, which become more sensitive to kanamycin and ampicillin. The genes encoding the AcrAB-TolC protein complex are a part of an operon, and a change in the DNA sequence of the acrA gene decreases the efflux of antibiotics. For example, AcrS is a protein that inhibits AcrA gene expression. The acrS gene extends 663 base pairs and is implanted in the acrEF operon [56]. AcrS attaches to the DNA promoter site of acrA required for initiating transcription, and then the bound AcrS inhibits the transcription of acrA.
Blocking the expression of AcrS increases antibiotic resistance in E. coli cells. After culturing E. coli in kanamycin at sub-inhibitory levels and deleting acrS in E. coli, increased levels of acrE transcription were exhibited. It was concluded that increasing AcrS expression increases the inhibition of acrE and this then decreases E. coli resistance to kanamycin [57, 58, 59, 60, 61, 62]. AcrS represses efflux pump structures and complexes. However, the effect of AcrS on controlling acrD mRNA transcription is not known [57, 58, 59, 60, 61, 62]. AcrD is an efflux pump for aminoglycosides related to other RND drug efflux pumps in E. coli. AcrD has been proven to induce adaptive resistance to kanamycin after culturing in kanamycin at sub-inhibitory levels [57, 58, 59, 60, 61, 62]. Cultures of E. coli, lacking acrS, showed more kanamycin resistance than the wild-type strain.
AcrS was determined to be an inhibitor of acrD expression. Therefore, inhibiting acrS can export more aminoglycosides through alternative and other RND efflux pumps [49, 50, 51, 52, 53, 54]. AcrD is a cytoplasmic membrane drug efflux exporter. AcrD is known to be a part of the generation of resistance to aminoglycoside antibiotics. Because AcrD is similar to AcrB, it is argued that AcrD forms a protein efflux complex with AcrA, which is a fusion protein located in the periplasm, and with TolC oriented in the outer membrane [63, 64, 65, 66, 67, 68]. After deleting acrD, the mutant acrD bacterial sample strain continued to develop antibiotic resistance and was not susceptible to four antibiotics when compared with the wild type strain [63-68]. As a consequence, further investigation of the effects of acrD is required. However, efflux pump blockers are increasingly toxic for human cells, but a nontoxic method of applying phosphorodiamidate morpholino rings can provide less toxicity via an antisense inhibition of antibiotic resistant mRNA transcription [56].
5. THE BINDING CAPACITY AND SPECIFICITY OF SUBSTRATES FOR ACRAB-TOLC EFFLUX
Four families of export pumps accompany the pumps classified as resistance-nodulation division (RND). The four other families are the major facilitator superfamily (MFS), the multidrop and TolC compound extrusion (MATE), the small multidrug resistance pumps (SMR), and the ATP-binding cassette (ABC) [69]. The ABC pump depends on the energy generated from ATP hydrolysis. RND pumps remove drugs and other toxic cations. RND pumps are a triplex of proteins that reside in the bacterial inner membrane region, in the outer membrane channel, and extend through the middle periplasm. The triplex releases drugs and toxins into the outer environment of the bacterial cell. After the toxins are removed, it is arduous for the antibiotics to regain re-entry into the cytosol through the bacterial outer membrane. The MFS and SMR pumps are less effective. The drugs are only secreted into the middle periplasm and not into the outer membrane by SMR and MFS pumps [70]. However, the RND pumps operate within the periplasm to expel many antibiotics remaining after MFS and SMR drug exportation. Because RND efflux pumps have a broad range of specificity for many types of substrates, this stagnates the discovery and design of novel antibiotic drugs [71, 72, 73, 74, 75, 76]. However, pathogenic bacteria must also overcome the oxidative activity of phagocytosis by immune cells. For pathogenic bacteria to survive phagocytosis and oxidative stress, the pathogenic bacteria increases RND pump expression to expel phagocytic oxidative compounds and particles from reactive oxygen species (ROS) and reactive nitrogen species (RNS) [77].
The substrates or antibiotics bind to the AcrB segment of the AcrAB-TolC protein pump. The nonpolar and highly hydrophobic substrates interact with the water molecules external to the bacterial cell. The antibiotics connect and form hydrogen bonds with the water molecules. The enzyme AcrAB-TolC binds the hydrophilic region of the antibiotics, which is then hydrogen-bonded to water [78]. The hydrophilic domains surrounding the antibiotics allow the facile removal and export of drugs from the bacteria. The multidrug pumps do not need specific residues or small enough binding sites for capturing and removing antibiotics [79]. Because of the water molecules forming hydrogen bonds with less stable antibiotic drugs, the binding site can be as large as possible for export, and this helps the pump overcome the initial threshold of binding free energy [79, 80]. Antibiotic resistance via efflux pump activity is observed only when the substrate is bound in the periplasmic cell region of the tripartite efflux pump. The substrate cannot remain in the cytosol of the bacterial cells [81, 82, 83, 84, 85]. For example, adding magnesium to AcrA rapidly increased the export of phospholipid-like substrates. The AcrA connects to two vesicles for transporting the phospholipids. Additionally, adding streptomycin, an antibiotic, to a more acidic mid-periplasm bacterial region of the AcrAB-TolC pump channel increased the frequency of pumping and efflux. Therefore, the removal of substrates or antibiotics can only occur via the AcrA subunit that is located in the periplasmic space [86].
The AcrB acts as a regulator when the concentration of antibiotics lessens the transmembrane pH gradient in the pump’s channel. The rate of hydrolysis in AcrAB-TolC affects the rate of efflux when calculating the Vmax and Km of enzymatic activity. The graphical analysis after comparing the rate of nitrocefin efflux to the periplasmic concentration of nitrocefin in the AcrAB-TolC pump yielded a Michaelis-Menten curve [81]. The Michaelis-Menten curve displayed a Vmax of 0.024 nmol/mg/s and a nitrocefin Km concentration of 5M, which demonstrated less competition and more cooperativity [81]. There was more cooperativity between the rate of efflux and the pump’s hydrolysis rate that was affected by the high concentration of nitrocefin. More antibiotics contained in bacteria lead to a higher velocity of efflux with rapid antibiotic removal, in which, the AcrB of E. coli increases the minimum inhibitory and minimal bactericidal concentration (MICs) of antibiotics [87]. The binding protomer of the AcrAB-TolC pump is mainly hydrophobic and lies centered within the bacterial periplasm. There are three different protomer activities of the AcrAB-TolC pump: 1) access, 2) binding, and 3) exportation. Each of the three subunit proteins of the AcrAB-TolC pump rotates as each protomer alters their orientation and conformations (Figure 2). The conformational changes are a result of the movements from the disulfide crosslinks and the adjacent side chain residue groups. The three subunits simultaneously rotate at least 1 or 2 subunits of the AcrAB-TolC protein complex at alternate times [81]. When antibiotics are present, the AcrAB complex begins to re-orient its conformation to allocate and recruit TolC [7]. The opening of TolC closes in the outer membrane to maintain the isolation of the periplasm from the external environment [7] (Figure 2). Then, the AcrAB-TolC pump opens by contracting in order to expel a substrate through its chamber and then rapidly closes after the expulsion of the drug molecule [7] (Figure 1).
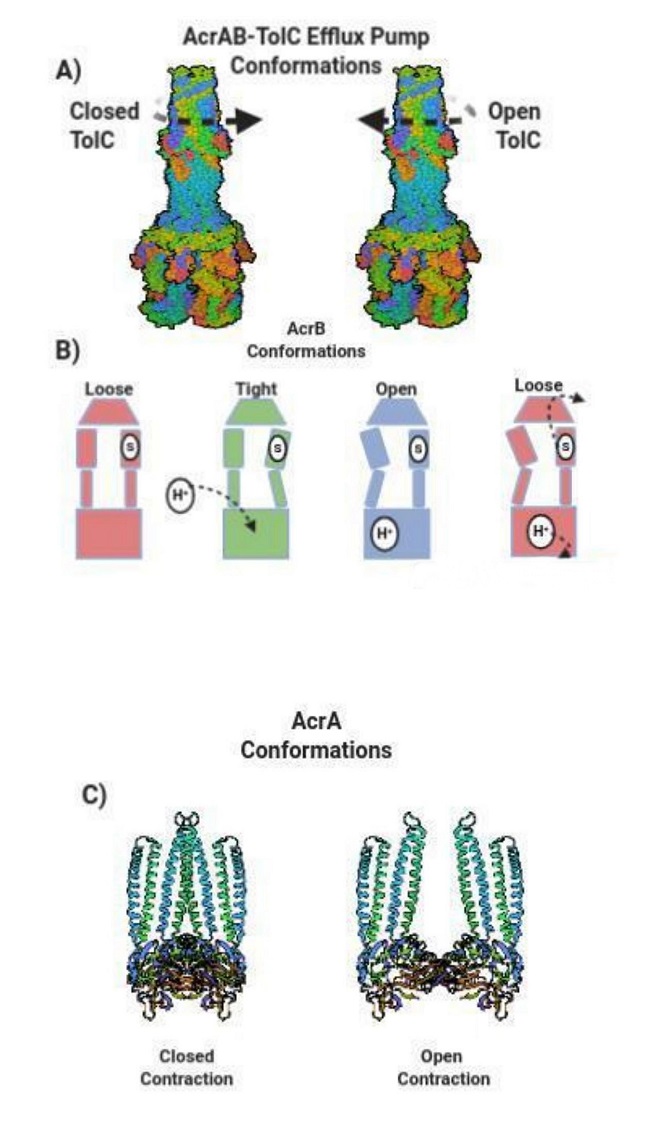
Figure 2: The Conformational changes of AcrAB-TolC Efflux Pumps. (a) TolC rotates counterclockwise and clockwise. Counter clockwise closes the TolC channel while clockwise rotation opens TolC. (b) AcrB conformations include loose, tight, and open orientations. A drug substrate (s) enters the initial loose form of AcrB and then changes into a tight position after a hydrogen proton enters AcrB. AcrB alters into an open adjustment and then returns to a loose assort-ment that forcefully exports the drug substrate. (c) AcrA contracts in a closed state and loosens with an opened contraction.
6. THE INHIBITION OF ACRAB-TOLC EFFLUX ACTIVITY
Li et al. reported the effects of site-directed mutagenesis of genes that encode the replacement of nonpolar phenylalanine R-groups within the AcrAB-TolC binding site [80]. Without phenylalanine, the efflux of antibiotics was blocked, and the phenylalanine was altered to express Phe610Ala. After the mutagenesis, the substrate and antibiotic doxorubicin remained bound to the binding site because the gene mutation of acrB caused the obstruction of the substrates’ separation from the binding site of the outer AcrAB-TolC pocket [88]. The acrB mutation caused the binding site to eliminate expression of the glycine loop, which removed the separation between the distal and proximal binding sites of AcrAB-TolC [88]. A mutation of acrB caused the efflux through the AcrB pump and channel to cease and this lessened the intensity of virulence. After the mutation of acrB, AcrB function is disrupted, and AcrB is aided by the transmembrane protein called D408. The D408 transports protons similar to AcrB. The mutation of the acrB gene incorporates a point mutation that affects the translation of the D408A transmembrane domain in AcrAB-TolC [89]. After the mutagenesis of the acrB gene, the movement of protons within the pump is decelerated and antibiotic efflux is inhibited. Salmonella Typhimurium SL1344 in mice, Galleria mellonella, and in tissue cultures all displayed less efflux following an acrB gene mutation [81]. The elimination of efflux for each organism was confirmed through RNA sequencing [89]. The D408A mutant also forced more regulation of gene expression for the flagella synthesis that is important for initiating the early stages of systemic infection.
The mutations of genes that encode the efflux pumps also inhibit the production of autoinducers (AI), which are quorum-sensing molecules, when there is less accumulation of AI molecules [89]. As a result, quorum sensing is not initiated, and virulent gene expression is decreased. Lux-S, which mediates and induces quorum sensing between bacterial cells, can be monitored for controlling the process of transcription. The upregulation of Lux-S causes metabolites to accumulate in the mutated bacterial strain where the high concentrations of metabolites triggers the upregulation of the EmrAB-MDR efflux pump [89]. EmrAB interacts with the TolC of the AcrAB-TolC pump, which removes antibiotics, toxins, and free fatty acids from bacteria. Therefore, inhibiting AcrAB-TolC activity can trigger EmrAB expression as a substitute, under pressure, and use available energy to remove the accumulated toxins. Nevertheless, the gene inhibition of acrB can increase antibiotic susceptibility; thus, Ayhan et al. also inhibited the acrA and tolC genes in E. coli.
In the Ayhan et al. study, inhibiting acrB, acrA, and tolC increased the levels of antibiotic sensitivity in E. coli. E. coli exhibited less antibiotic resistance after inhibiting these three genes [30]. Ayhan et al. targeted the mRNA for each of the three genes with PPMOs. The PPMO of acrA produced the most impactful antisense molecules where E. coli treated with acrA-PPMOs increased the antibiotic effectiveness from 2 to 40-fold [33]. The AcrA protein was not further translated, and the efflux of antibiotics was diminished without any toxicity to human cells [33]. The inhibition of acrA may have yielded the most significant reduction of antibiotic resistance because the AcrA protein subunit mediates communication between AcrB and the TolC for the purpose of controlling the pump’s closure and opening [7]. The AcrA hexamer is pertinent because it guides the opening of the TolC, which then allows the construction and activation of the AcrAB-TolC tripartite complex [7]. Thus, inhibiting the genes that code for efflux pump protein synthesis can reduce the expression of efflux pumps and decrease antibiotic expulsion. Inhibiting the expression of MDR efflux protein pumps can increase the bacterial sensitivity of antibiotics.
7. BACTERIAL METABOLISM AND ANTIBIOTIC RESISTANCE
Stimulating the metabolic pathways of bacteria can increase drug uptake in antibiotic tolerant bacteria [90, 91, 92]. For example, cellular respiration regulates tobramycin antibiotic potency by increasing drug import and through elevating bacterial cell death. The citric acid cycle (TCA cycle) intermediate of fumarate can amplify tobramycin susceptibility [90]. Researchers also cultured alanine and glucose with the bacteria known as Edwardsiella tarda, which is a bacterium resistant to multiple antibiotics such as tetracycline, chloramphenicol, streptomycin, and sulfonamide. They examined the potential of alanine and glucose to increase the kanamycin sensitivity of E. tarda. Their predictions and hypotheses were confirmed, adding alanine and glucose with kanamycin to the E. tarda bacterial cultures eliminated many bacteria. The E. tarda cultured with only 1000mg of kanamycin, 40mM of alanine, and with 10mM of glucose decreased the number of bacterial cells by 101, to 3228, and then to 276,000-fold, respectively [93]. Therefore, it has been concluded that glucose elevates the uptake of antibiotics by increasing the activity of the proton motive force or PMF. Glucose becomes converted into pyruvate and vice versa via the glycolysis/gluconeogenesis pathway. Metabolism of pyruvate is the final stage of the TCA cycle that increases NADH levels, which then amplifies PMF. The increased PMF can increase the antibiotic uptake of aminoglycosides [94, 95]
Certain metabolic stimuli can induce aminoglycoside eradication of Gram-negative and Gram-positive persisters. For example, after activation and amplification of PMF, the uptake of aminoglycosides is advanced and elevated [96-98]. Culturing E. tarda with alanine and/or glucose resulted in E. tarda cell induced apoptosis [93]. Pyruvate increased proportionally to the elevated levels of alanine and/or glucose [93]. The concentrations of NADH and PMF also were amplified. It was concluded that alanine and glucose increased and induced the absorption of antibiotics [85]. The kanamycin inside of the E. tarda cells were in high concentration after exposure to alanine and glucose. Bacterial samples without alanine or glucose exhibited a kanamycin uptake of 9.5 ng/mL, however, the cultures with alanine and glucose raised the absorption of kanamycin to 65-123 ng/ml and then to 113-231 ng/ml [93]. The alanine and glucose cultures of E. tarda trounced the activity of the multidrug efflux pump removal of antibiotics.
Stimulating the metabolism of E. coli cells can also increase its uptake of antibiotics. Through a Michaelis-Menten kinetics experiment, researchers found E. coli K12 to develop an enhanced sensitivity to the gentamicin antibiotic when cultured in Luria-Bertani broth supplemented with glutamate and acetate [99]. The cycle of NADH plus PMF was significantly increased after culturing E. coli cells with glutamate [99]. The increase in the gene expression of maeA and maeB produced more pyruvate in the presence of glucose, which indicates that the TCA cycle is dependent on the pyruvate cycle, termed the P-cycle [88]. Manipulating the P-cycle through the OAA-PEP-pyruvate-AcCoACIT pathway and the TCA cycle may be a favorable target for inhibiting antibiotic resistant genes [96]. The P-cycle depends on the abundance of exogenous metabolites, so as a result, antibiotic resistance to aminoglycosides could continue to decrease after genes of the P-cycle are also silenced [96]. In striking contrast, bacteria rapidly become more resistant to antibiotics when sources of carbon and nutrients are less available. For instance, bacterial species detect and respond to the unavailability and limitation of nutrients via a regulatory mechanism termed the stringent response (SR) [100]. Less access or starvation of carbon, amino acids, and iron stimulates the SR by activating the expression of relA and spoT gene components to construct the alarmone known as (p)ppGpp. SR was inactivated by inhibiting relA and spoT gene expression in Pseudomonas aeruginosa that causes infections and is used as a model to study biofilm formation. After inactivating SR, (p)ppGpp synthesis was eliminated through the starvation-induced serine analog called serine hydroxamate (SHX) [100].
8. BACTERIAL METABOLISM AND THE ACRAB-TOLC EFFLUX PUMP
Additionally, when the AcrAB-TolC pump is inactivated or inhibited, metabolites begin to accumulate that then inactivate AcrR. Ruiz and Levy reported that the accumulation of toxic metabolites affect the expression of transcriptional controllers of acrAB such as AcrR [93]. Thus, Ruiz and Levy confirmed that the AcrAB-TolC pump controls the expression of the acrAB operon by responding to changes in cellular metabolism [101]. For example, AcrAB-TolC efflux is dependent on the PMF acquired through the electron transport chain of bacterial metabolism and generated by the electro-chemical gradient of protons formed across the cell membrane. AcrB is a component of the multidrug efflux pump that functions as a drug/proton transport system [102]. The PMF provides energy for multidrug resistant AcrAB-TolC efflux pumps. The functions of PMF include coupling membrane-associated enzymes, transporting solutes through the membrane, or maintaining the pH of the cytoplasm [103]. During the activity of PMF, protons flow inwardly into the cytoplasm that energizes efflux activity [102]. A proton is transferred from the periplasmic space and pool to the D407-D408 pair of AcrB, and then the positive charge of a side chain in the AcrB protomer returns to the D407-D408 pair for deprotonation. After the proton is released, the proton continues to cross the membrane, and then the remaining energy from PMF is released [63, 64, 65, 66, 67, 68]. Inhibiting efflux activity and increasing antibiotic susceptibility are possible through altering the levels of PMF in the AcrB protomer. Using plating assays revealed that these acrAB mutants were more susceptible to polymyxin B by 10,000-fold [103].
Additionally, treating the E. coli acrAB mutant cultures with the PMF uncoupler termed carbonyl cyanide m-chlorophenyl hydrazone or CCCP increased polymyxin susceptibility [104]. A possible explanation for the increased antibiotic susceptibility of acrAB mutants after CCCP treatment includes that a lesser frequency of protonation and deprotonation in AcrB lowers the PMF activity needed for powering AcrAB-TolC efflux. For instance, the three amino acid residues contained in AcrB such as the acidic Asp407 and Asp408 residues, located in the transmembrane (TM) helix termed TM4, with one basic side chain residue of Lys940 in TM10, form salt bridges and hydrogen bonds (H-bonds) that participate in the transfer and transport of protons [102]. Because AcrB consists of two acidic residues with one basic side chain residue, replacing one of these residues with an alanine, within the D407-K940-T978-D408 complex of AcrB, suppresses the salt-bridge and H-bonding interactions [99]. The salt bridges and H-bonds are perturbed because the alanine cannot become protonated or deprotonated [99]. Mutations of acrB reduce protonation and deprotonation reactions required for triggering the PMF essential for AcrAB-TolC efflux [99]. In addition, by blocking the cellular assembly of protein efflux pumps, the antibiotic transport through the protein pump channel can become reduced and impede the energy garnered through ATP hydrolysis required for powering pump activity. However, currently, the effects of PMF uncouplers and blockers on MDR efflux pump activity are not sufficiently understood [104].
9. CONCLUSIONS AND PERSPECTIVES
Developing novel antibiotics to combat bacterial antibiotic resistance has become stagnant because of many economic and governmental obstacles. In 2013, according to the Infectious Diseases Society of America (IDSA), not many antibacterial agents passed into phase 2 or phase 3 of development [105]. Approximately, 15 of 18 pharmaceutical companies have relinquished developing antibiotics [105]. Academia has also dwindled its discovery of new antibiotics due to less availability of funding [105]. The development and discovery of new antibiotics is presently not considered a savvy economic investment in the pharmaceutical industry [105]. Since antibiotics can quickly cure infections, antibiotics are not as expensive and cannot produce a profit as amply large as drugs used for chronic conditions such as asthma or diabetes. Pharmaceutical companies have a greater propensity to invest in developing medicines used for chronic conditions because these types of medicines are immensely profitable [105]. An additional factor that has lessened the development of antibiotics includes a deficiency of financial attraction because antibiotics are sold at lower prices. Antibiotics that are new have a price of 1,000 USD to 3,000 USD that is significantly lower than the 10,000 dollars or more drugs used for cancer chemotherapy [105]. Additionally, many microbiologists have argued for lesser use of antibiotics and this has caused many physicians to reduce their prescribing of many antibiotics in fear of their proliferation of bacterial drug resistance [105].
Therefore, because there is an urgent need to develop new antibiotics, this review attempted to provide an alternative perspective in support of discovering novel antimicrobials that are gene-based. In this review, an alternative antimicrobial design was discussed that can target the genes responsible for expressing specific subcellular and molecular components, which may induce the antibiotic sensitivity of multidrug resistant bacteria. The susceptibility of bacteria to develop increased sensitivity of antibiotics depends on a few conditions, such as the bacterial metabolism of carbon, exposure to metabolites, its horizontal transfer of antibiotic resistant genes, efflux pumps, and a bacterium’s direct hydrolysis of antibiotic drugs. In this review, it was noted that a mutated acrB gene could alter the AcrAB-TolC binding site by substituting a nonpolar phenylalanine amino acid residue with an alanine. The mutagenesis of the acrB gene, encoding the efflux pumps’ binding sites for AcrAB-TolC, inhibited the exit of antibiotics through the AcrAB-TolC efflux pumps. Gene inhibition of the acrA and the acrB genes in E. coli lowered the level of virulence and increased antibiotic efficacy. Moreover, altering the metabolic output of an MDR bacterial cell by exposure to glucose increased the antibiotic sensitivity of E. tarda. Researchers cultured E. tarda with glucose and alanine, and the uptake of kanamycin increased, eliminating approximately 3,000 times the number of MDR bacteria compared to the cells only treated with kanamycin. A mutation or deletion of acrB can reduce the levels of protonation and deprotonation, which then limits the energy produced by PMF for AcrAB-TolC efflux.
Novel synthetic biological strategies can be used for re-engineering complex microbial pathways and networks, which is currently becoming more simplistic and increasingly practical [106]. Accordingly, through re-engineering plasmids, bacterial gene circuits may be rewired to upregulate metabolic genes and inhibit genes that encode efflux protein pump expression. Plasmids can become genetically engineered to induce transcription of metabolic genes such as maeA and maeB, thereby, amplifying the pyruvate production that increases PMF. The higher levels of PMF activity intensifies antibiotic uptake. Simultaneously, plasmids could be synthetically engineered to produce the clustered regularly interspaced short palindromic repeats and CRISPR-associated protein 9 (CRISPR-Cas9) nuclease deletion of genes such as acrA and acrB or small interfering RNAs of these genes can reduce MDR drug efflux. The advantage of the CRISPR-Cas system is that it offers more weapons of warfare against antibiotic resistant pathogens. Research scientists using the CRISPR-Cas system can find alternative solutions for extracellular infections such as MRSA and other intracellular antibiotic resistant bacteria such as Burkholderia pseudomallei. However, presently, there remains an intense challenge when applying CRISPR-Cas9 antibacterials against resistant bacteria external to the laboratory because there may exist many ethical concerns for changing or modifying bacterial genomes. Additionally, the delivery of antimicrobial gene therapies is presently a major challenge [107]. Bacterial metabolism of multi-drug resistant bacteria can alter efflux pump activity. Moreover, stagnating efflux pump drug export can affect the metabolism of drug-resistant bacteria. Perhaps, targeting the genes that regulate bacterial metabolism and generate efflux pump activity may reduce antibiotic resistance.














