INTRODUCTION
Plasmids are external genetic components that can become mobile. Plasmids can become transferred between bacterial cells via conjugation. Plasmids carry and transport genes that assist their hosts to survive in a plethora of environments [1]. The ability of plasmids to carry and transfer genes can initiate and complete the evolution and ecology of bacterial cells [1]. Because plasmids disseminate antibiotic resistance genes, these antibiotic resistance modes of action can continue to persist among medical pathogens for many additional decades [2]. These plasmids act as a type of genetic parasite [1]. Antibiotic resistance genes serve as benefits encoded in plasmids that can lead to virulence, high tolerance of heavy metals, and to higher rates of carbon metabolism [1]. Plasmids can also alter the environment of surrounding bacterial cells where beneficial enzymes or sources of nutrients can be produced [1]. Although plasmids are found in bacterial populations, plasmids can form many changes of physiology in their bacterial hosts, which then reduces plasmid fitness [3]. The cost to fitness has created a quandary and a difficulty for interpreting how plasmids are conserved in a bacterial population for the long-term [4]. The challenges of understanding plasmid persistence has led to an unsolved riddle known as the “the plasmid paradox” [4].
Many hypotheses have been presented to attempt to solve this paradox. For example, it has been argued that bacterial evolution fuels plasmid persistence by lessening the fitness cost combined with plasmid carriage where an amplified rate of conjugation solidifies the persistence and survival of plasmids [5]. As a result, the persistence and containment of plasmids in bacterial populations over time presents to the current field a plasmid persistence puzzle or a plasmid paradox. In theory, plasmids carrying beneficial genes do not provide a solid explanation for the maintenance of plasmids [6]. Because of fitness costs, plasmids in theory should dwindle while the beneficial genes integrate within the chromosomes of bacteria. As a result, many additional hypotheses have been generated to more succinctly explain the paradox of plasmid persistence. These hypotheses include a higher frequency of HGT and conjugation during infectious transmission, the plasmid transport between strains of bacteria, and in the maintenance of the higher proliferating bacterial strains, which display amplified effectiveness of transconjugation [6].
For many decades, a large number of studies have examined the conditions for plasmid persistence and maintenance in bacterial populations. A limitation takes into consideration that a majority of the mathematical models for examining plasmid population biology research involves bacterial populations within similar colonies. However, most bacterial cells inhabit complex communities where plasmids disseminate through conjugation from varied bacterial hosts [1]. Therefore, there is a gap in knowledge concerning the persistence of plasmids in natural bacterial populations and communities [1, 2]. It is crucial to resolve these limitations to enhance our understanding of plasmid persistence. To address a few of these gaps in knowledge, the present study attempted to examine plasmid persistence in relation to plasmid carriage in Bacillus subtilis co-cultures. B. subtilis inhabits soils and dwells in many environments and ecosystems. B. subtilis are also gut commensal bacteria found in animals and in humans [7]. B. subtilis was selected for this study because it uses plasmids to assist with HGT in the gut microbiome [7]. Moreover, more frequent HGT events occur in the human gut microbiome than in a diverse ecosystem of non-human microbes [8]. Physical proximity contributes the most to the large operation of HGT in the gut via cross-domain gene transfer [9]. Conjugation as a mechanism of HGT is substantially altered by physical proximity [9]. Conjugation is effective in the gut because it conserves and protects DNA from enzymes known as nucleases along with elimination of heavy metals in the environment [9]. Conjugation via HGT has a highly significant role in the gut microbiome, and an improved understanding of conjugation's contribution to HGT in the gut is greatly needed [9].
Therefore, the main objective of this study was to provide additional evidence and explanations for plasmid persistence. To offer a potential interpretation of plasmid persistence, a possible relationship between natural bacterial conditions and plasmid persistence were investigated in B. subtilis (B. sub), Vibrio harveyi (VH), and Escherichia coli .E. coli) bacterial cell co-cultures. These bacterial strains were selected to model in vitro and naturally occurring communities of bacteria that represent an interaction between a few bacterial phylotypes present in the gut microbiome. For example, in this study, B. subtilis and E. coli that inhabit the gut microbiomes of animals and humans were used while V. harveyi was examined, which is a pathogen found in fish, in which many humans and animals consume. Factors and conditions, such as bacterial growth, biofilm formation, and antibiotic resistance genes, which may contribute to plasmid persistence, were analyzed in this study. These natural phases of bacterial populations were observed for any changes in bacterial host fitness as related to plasmid carriage that may offer insights into the dynamics and contributors of plasmid persistence. It was hypothesized that plasmid carriage can alter the natural phases of bacterial communities, which all interrelate with the plasmid persistence of bacterial host cells. To test the hypothesis of this study, B. subtilis cells were transformed with Clustered Regularly Interspaced Short Palindromic Repeats or CRISPR plasmid vectors encoding human CRISPR associated protein 9 (hCas9) of accA, which codes for the bacterial acetyl-CoA carboxylase subunit AccA. The plasmid utilized a promoter exclusive only for human RNA polymerase to bind so that the bacterial accA-sgRNA could not be expressed. This was done in order to only observe the effects of plasmid carriage without gene modifications in the target bacterial host. After transformation of B. subtilis with the CRISPR-hCas9-accA plasmid vectors, the B. subtilis plasmid carriers and non-carriers (B.sub-C/B.sub-NC) were co-cultured with E. coli and V. harveyi (VH) in order to exhibit any changes in the conditions of the bacterial cell co-cultures. The bacterial cell growths, biofilm intensity, and the antibiotic resistance of the co-cultures were then examined via common microbiological assays such as a biofilm crystal violet assay. As a consequence, the purpose for this study was to observe and analyze possible significant changes in the innate phases of bacterial communities to find a significant link between the three aforementioned natural bacterial states and the plasmid carriage of the bacterial co-cultures, which all can maintain plasmid persistence.
MATERIAL AND METHODS
BACTERIAL STRAINS
Bacterial strains included B. subtilis, V. harveyi DSM 6904 (DSMZ), E. coli BL21, and E. coli DH5a. Luria Bertani broth and agar were inoculated with each strain of bacteria aforementioned. The LB liquid and LB agar media were incubated from 16 hours to 24 hours at the temperatures specific for each bacterium. B. subtilis and V. harveyi bacterial cells were incubated at 30 degrees Celsius as E. coli cell cultures were grown at 35 to 37oC.
PLASMID DESIGN AND ASSEMBL
The vector named pRP[CRISPR]-hCas9-U6>{accA} was designed in VectorBuilder.com. The vector ID is VB210702-1210yff with a total value of 8431 base pairs (bp). The vector was designed to contain a single guide RNA (gRNA) for CRISPR-hCas9 to carry the 20bp of the accA gene. The gRNA can express the sequence of CTGCGTGAAATGTCTCGCCT, which is a guide RNA of 20bp for the bacterial accA gene. To assemble pRP[CRISPR]-hCas9-U6>{accA} hCas9 was inserted into the vector. The antibiotic resistant gene for ampicillin was also included in the plasmid assembly. The plasmids were produced at a high plasmid copy number. The fully assembled vectors were assembled and delivered by VectorBuilder. The plasmids were delivered through a glycerol stock solution of E. coli cells. The pRP[CRISPR]-hCas9-U6>{accA} plasmids were purified from the E. coli cells via the BioBasic EZ-10 Spin Column plasmid DNA Kit (BS413-50 preps).
B. SUBTITLIS TRANSFORMATION
The pRP[CRISPR]-hCas9-U6>{accA} plasmids isolated from the E. coli stock of cells were used to transform B. subtilis cells, deriving the B. subtilis-Carriers. To optimize the B. subtilis transformation protocol, B. subtilis was cultured for 5 hours until the exponential phase of growth with an OD600 value of 0.3-0.4. Two millimeters were taken from the culture and centrifuged for 1 minute at 8,000 rpm, collecting the B. subtilis cells. The supernatant was discarded. About 200 μL of LB broth with 2% xylose was added and then vortexed to concentrate the cell suspension at 10x. The 2% xylose was added to the LB. The isolated pRP[CRISPR]-hCas9-U6>{accA} plasmids with ampicillin selective markers were added to the concentrated B. subtilis cells with the 2% xylose-LB solution. The B. subtilis transformations were incubated for 1 hour at 37o C and then plated. About 20 μL each of the pRP[CRISPR]-hCas9-U6>{accA} B. subtilis transformations were inoculated onto two LB agar plates and 160 μL were also plated. The plates were sealed with parafilm and incubated overnight at 37oC. Each LB agar culture plate contained carbenicillin to select for the pRP[CRISPR]-hCas9-U6>{accA} plasmids with the ampicillin selective marker because carbenicillin cannot degrade in high acidity or in excessive heat.
CO-CULTURE ASSAYS
The V. harveyi and the E. coli bacterial cells were co-cultured with the B. subtilis Carriers and the wild type (WT) B. subtilis to detect any change in growth or bacterial host fitness of the B. subtilis-Carriers. After completing the co-cultures, the B. subtilis-Carrier co-cultures with V. Harveyi and E. coli BL21/DH5α cells were compared to the co-cultures of the WT B. subtilis with the V. harveyi and the E. coli BL21/DH5α. To use a co-culture assay and approach, each bacterial species and strain, aforementioned, were subcultured on seperate agar plates. Colonies from the agar plates were inoculated into LB liquid medium and then incubated overnight. About 5-6 bacterial colonies from each E. coli BL21/DH5α and V. harveyi plate and 5-6 colonies from the B. subtilis-Carrier and WT B. subtilis agar plates were inoculated into LB broth. The co-cultures without the B. subtilis carriers were used as the control samples. The co-cultures were grown overnight at 30 to 35oC and then analyzed for CFU/mL, biofilm production, and antibiotic resistance.
COLONY FORMING UNITS (CFU)
To measure the total of colony forming units (CFU), 10μL of each stock of the bacterial strains were pipetted into 1.5 mL Eppendorf centrifuge tubes. About 0.1mL of the 5 mL stock was added to 0.09 mL of LB broth to form the 10-1 dilution of each bacterial strain. The dilutions were resuspended. Then, 10μL of the 10-1 dilution was added to 90μL of LB broth to form a dilution of 10-2. The 10μL of 10-2 were added to 90μL of LB broth, and then more dilutions were continued in a series to a total of 10-10 dilutions. The last dilution and two other dilution series were selected for plating. The plates with the selected dilutions were incubated overnight at 37oC. The number of colonies for each bacterial strain was counted from the cultured agar plates, using the Promega Colony Counter computer software program. The number of colonies counted was substituted into an equation for calculating the values of CFU for bacterial cells.
The equation included:
CFU/mL=[Number of Colonies Counted] x [Dilution Factor]/Volume spread on plate
KIRBY-BAUER DISK DIFFUSION TESTS
The co-cultures of E. coli BL21/DH5α and V. harveyi with B. subtilis Carriers and WT B. subtilis were then assayed for antibiotic resistance. The Kirby-Bauer Disk Diffusion Tests were used. Carbinicillin, chloramphenicol, tetracycline, and kanamycin were pipetted into sterilized antibiotic disks. Approximately, 5 μL of each of the four antibiotics were pipetted into the antibiotic disks. A lawn of each bacterial strain was inoculated on LB agar plates via the spread plate method. A swab of LB broth liquid cultures from each bacterial strain was spread onto agar culturing media. The antibiotic disks were placed onto the bacterial culture plates. The plates were incubated for 24 hrs at 35oC. The zones of inhibition were measured in millimeters (mm) and then the values were interpreted, using a zone diameter interpretive standards chart provided by Sarker et al. [10].
BIOFILM ASSAY
Five milliliters of LB broth cultures were prepared from each bacterial strain and co-culture. The biofilms of the E. coli and V. harveyi strains were used as the controls and were co-cultured with the B. subtilis Carriers and WT B. subtilis. The cultures grew for 20 hours at 37oC. The 1: 100 dilution sets were prepared, totalling to 1mL of the LB/Carb liquid cultures. About 100 μLof the dilution series were pipetted into 4 wells, per bacterial strain, of a 96 well plate. The plates were incubated for 48 hours at 37oC. The plates were shaken to remove any planktonic bacteria. The plates were rinsed with water. All the wells were stained with 125 μL of 0.1% crystal violet solution for 10 min. The 96-well plate was shaken and rinsed with water to remove any excess crystal violet solution.
STATISTICAL ANALYSIS
The p-Values of Colony Forming Units for V. harveyi samples were analyzed through a Paired-Two-Tailed t-test. The biofilm assays were analyzed via Two-Way ANOVA. The antibiotic test data results were analyzed by One-Way ANOVA for E. coli samples as paired-t-tests, which were used to generate p-Values for V. harveyi antibiotic resistance sample results. Descriptive and inferential statistics were performed, using Microsoft Excel and GraphPad Prism. All graphs and other statistical analyses were generated through ChartExpo and GraphPad Prism.
RESULTS
BACTERIAL GROWTH DECREASED IN V. HARVEYI, E.COLI, AND B. SUBTITLIS-CARRIER CO-CULTURES
After transforming B. subtilis cells with the CRISPR-hcas9-sgaccA plasmid vectors, the transformants were co-cultured with V. harveyi and E. coli. The B. subtilis cells that were transformed with CRISPR-hCas-9 plasmid vectors were compared against bacterial samples with non-carriers such as the V. harveyi and the E. coli. The colony forming unit (CFU) values of the non-carriers were compared to the carriers. As a result, after calculating the CFU/mL, bacterial growth, in the B. subtilis-Carrier and V. harveyi co-cultures, significantly decreased with a paired-t-test two-tailed P=0 (Figure 1A). V. harveyi decreased by an average of 31% after the co-culture assays (Figure 1A). The bacterial growth between E. coli and B. subtilis Carrier co-cultures produced a percent difference of 54% (95% CI [3.06M, 1.10M]). E. coli yielded 3.02E7 CFU as the co-culture decreased to 1.10E7 CFU. V. harveyi had an average CFU of 5.8E11 that was then reduced to 3.4E11 CFU during the co-culture assays (Figure 1A). Overall, the co-cultures containing B. subtilis plasmid carriers exhibited lesser bacterial growth (Figure 1B).

Figure 1. The Colony Forming Units (CFU/mL) After dilution series for each sample were generated, the CFU/mL were allocated. Each dilution series showed a large decrease in CFU/mL for B.subtilis-Carrier-V.H compared against the V. harveyi only bacterial sample culture.A. A significant decrease in bacterial growth occurred for the B.subtilis-Carrier-V.H-E.Coli co-culture assays.B. V.H CFU/mL dwindled from 3.69B to 2.46B when V. harveyi was co-cultured with B.subtilis plasmid carriers. E. Coli, when co-cultivated with plasmid carrier B.subtilis, displayed a significant decrease in CFU/mL from 3.02M to 1.10M.
BIOFILM FORMATION WAS AMPLIFIED IN B. SUBTITLIS CARRIER-VH CO-CULTURES, BUT REDUCED IN E. COLI CO-CULTURES
Biofilms include populations of microorganisms growing on a surface within a matrix of extracellular polymeric molecules. An innate property of biofilms is that biofilms are spatial and organized shelters where microbes interact with adjacent microbial communities rather than with wider spaced bacteria. The spatial structural complex offers heterogeneity within these spatial structures of biofilm where bacteria experience different environmental conditions and pressures in different locations of the biofilm [11]. The spatial structure and the heterogeneity of this biofilm habitat significantly affect the evolution of populations of microbes [11]. For example, through slowing down competition between bacterial cells, biofilm spatial structures' environments select for many favorable mutations and then delay the rate of adaptation [11]. Thus, the biofilm intensity of each sample, including WT--VH-E.coli, B.sub-C-VH-E.coli, and WTB.Sub-NC-VH-E.coli, was measured through a crystal violet assay. Biofilm formation was amplified in B. subtilis Carrier-V.H co-cultures, but reduced in the E. coli co-cultures. Biofilm is vital for the survival, persistence, and for the proliferation of bacterial cells. Biofilm serves to insulate and protect bacterial cells from external antimicrobials such as antibiotics and other antagonistic microbes. Biofilm is also a major contributor to the increased virulence and proliferation of pathogenic bacteria, and it also invigorates antibiotic resistance. The biofilm production of V. harveyi and E. coli co-cultures with wild type B. subtilis were compared to the B. subtilis plasmid carriers, V. harveyi, and E. coli co-cultures. The wild type bacterial strains of V. harveyi and E. coli were used as control samples for comparative analysis. For V. harveyi samples grown with B. subtilis Carriers, biofilm formation exceeded the levels of biofilm generated by V. harveyi combined with WT B. subtilis. A Likert scale ranging from 1 to 5 was used to quantify the intensity of crystal violet absorbance added to the samples. A score of 4-5 equaled a high intensity of crystal violet coloration, and a value of 3 to 4 represented a moderate intensity of crystal violet absorbance. If a sampling well did not appear with much coloration or no stain was formed, these samples were given a score of 0 to 1.
The B. subtilis-C-V.H co-cultures averaged a score of 3.9, and the WT B. subtilis-V.H samples averaged a 3.3 crystal violet (CV) assay score. The control of the WT V. harveyi averaged a 3.8 CV intensity score. WT B. subtilis-V.H samples, the B. subtilis Carrier-V.H co-cultures, and the controls each scored a total of 40, 47, and 46 of CV intensity, respectively (Figure 2A-2B). The p-Values, after performing a Two-Way ANOVA analysis of each group's CV scores, computed to P=0.134, P<.001, and P=0.64 for comparing the difference between the scores, the difference between the bacterial sampling groups, and for determining the variable difference between the bacterial groups and their CV scores, respectively. There was an increase in biofilm yield when comparing the B. subtilis-C-V.H treatment groups against the WT B. subtilis-V.H sample groups. However, the CV scores, for the B. subtilis-C-V.H co-cultures, of biofilm formed were equally approximate to the WT-V.H sample scores. Biofilm formation decreased after co-culturing E. coli with B. subtilis-Carrier, yielding a P<0.001 (Figure 3A-3C).
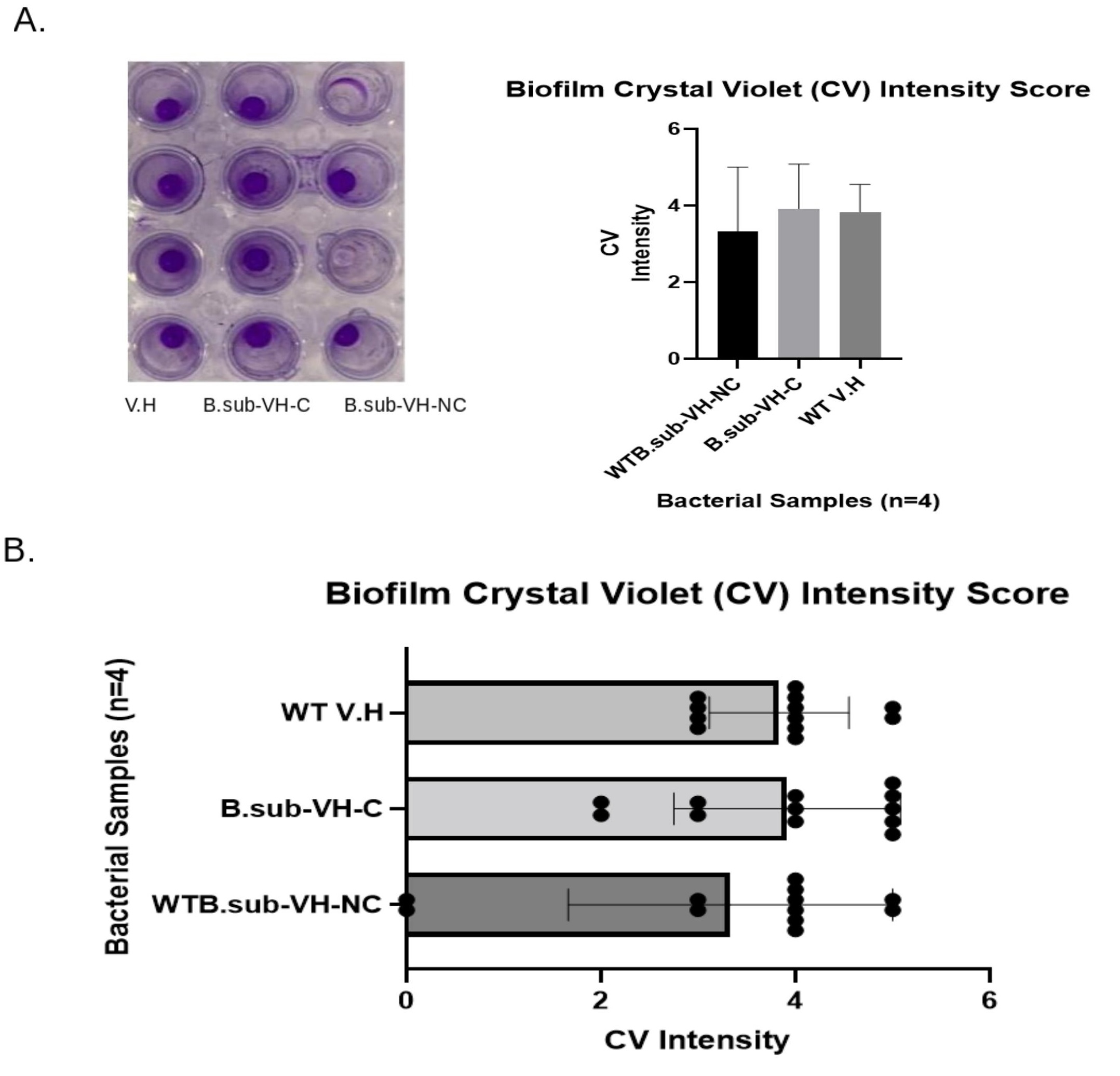
Figure 2. Biofilm Assay.A. The crystal violet stain of B. subtilis-Carrier-V.H appeared with the highest intensity. B. subtilis-Carrier-V.H and V.H showed a higher score of CV intensity than the WT-B. subtilis-V.H co-cultures. B. The biofilm assays were performed in triplicates, in which the B. subtilis-Carrier-V.H had the highest expression of CV absorbance intensity. B. subtilis-Carrier-V.H co-cultures showed a higher intensity of Crystal Violet absorbance versus the wild type B. subtilis as shown in the simulation of data and in the scatter plot.
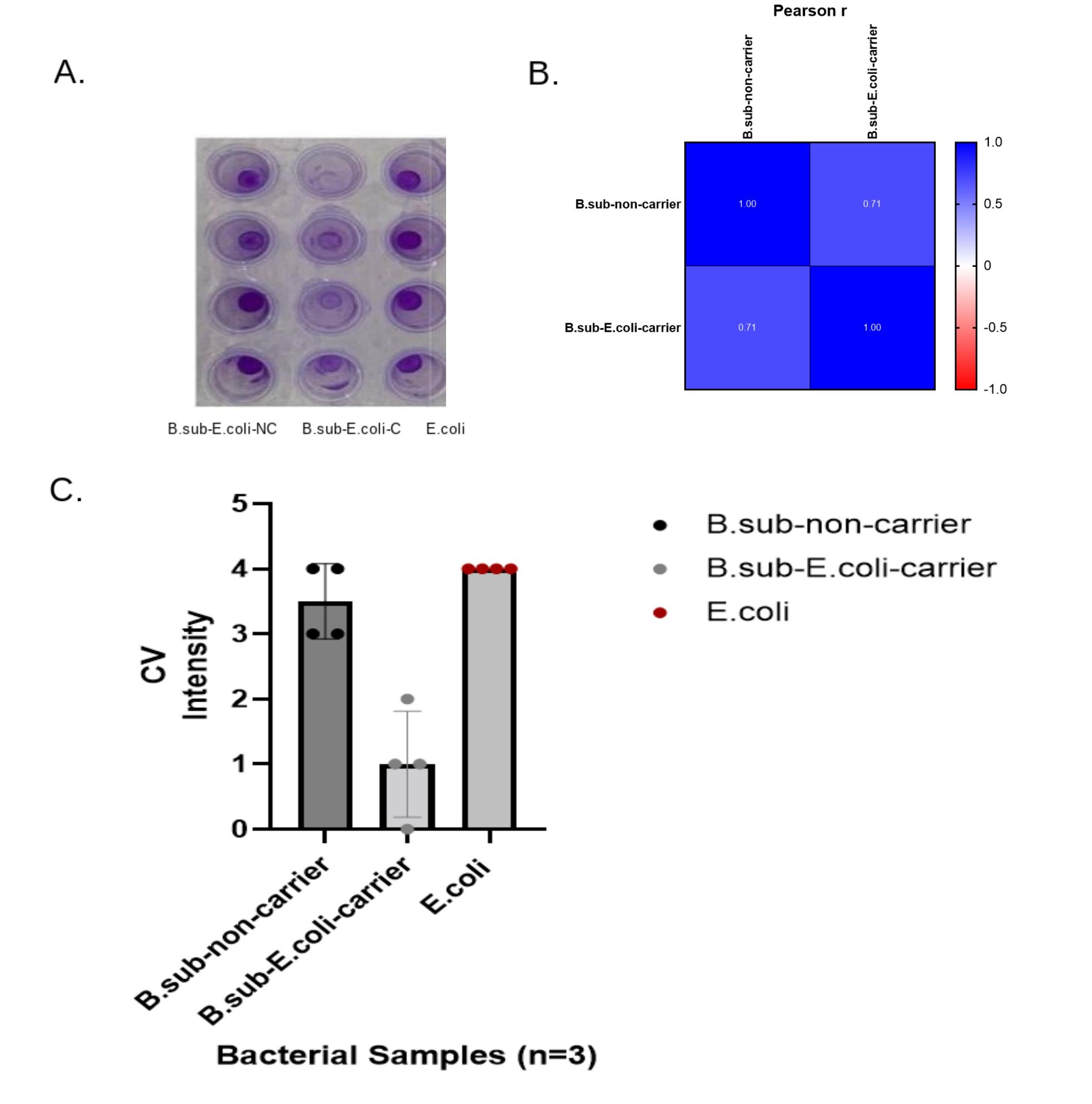
Figure 3. Biofilm Assay for E. Coli and AB. Subtilis Carrier.A. E. Coli had a total score of 16 with an average CV intensity of 4 (Avg=4, SD=0). B. The Pearson r analysis shows coefficients of 1 between the non-carriers while there was a coefficient of 1 in the co-culture samples of carriers. There was a 0.71 coefficient between the noncarriers and the carriers, showing a decrease in correlations between each sample. C. B.subtilis-Carriers combined with E. Coli averaged a score of 1 CV intensity and a total score of 4 (Avg.=1 SD=0.8). E. Coli, grown with WT B. subtilis, averaged a 3.5 CV score and had a total score of 14 (Avg=3.5, SD=0.58).
B. SUBTITLIS CARRIER-V.H AND B. SUBTITLIS CARRIER-E. COLI CO-CULTURES SHOWED MORE ANTIBIOTIC RESISTANCE THAN CONTROL SAMPLES
The Kirby-Bauer antibiotic disk assay was used to analyze the antibiotic resistance of B. subtilis-Carrier co-cultures of B.sub-C-VH-E.coli and the non-carriers such as the B.sub-NC-VH--E.coli, including the control samples of WT-VH and WT-E.coli. The antibiotic resistance of B. subtilis-Carrier-V.H and the V. harveyi cultures were closely identical (Figure 4A). In fact, the B. sub-C-V.H cultures seemed to share the exact amount of antibiotic resistance with nearly equal zones of inhibition in millimeters. All co-cultures were resistant to carbenicillin and moderately resistant to tetracycline (Figure 4A). The B. sub-C-V.H, the WT-V.H, and the control samples were all sensitive to chloramphenicol and to kanamycin. The antibiotic resistance of B. sub-C-V.H co-cultures compared against the control samples had a Two-Tailed P=0.51 after using a paired t-test analysis. The E. coli samples showed similar levels of antibiotic resistance as the B. sub-C-E. coli co-cultures. All B. sub-C-E. coli sample groups were more resistant to each antibiotic. The B. subtilis-C-E.Coli compared against E. Coli, after a One-Way ANOVA analysis, yielded a P=0.49. The antibiotic resistance levels of the co-cultures seemed to mimic the antibiotic resistance expressed by the control sample groups, which included V. harveyi or E. Coli; however, the antibiotic resistance increased by 3% for the B. sub-C-V.H samples while the B. sub-C-E. coli co-cultures decreased in antibiotic sensitivity by approximately 1.5% (Figure 4B). The B. subtilis carriers co-cultured with V. harveyi and E. coli displayed a higher level of antibiotic resistance than the controls and its wild-type because the B. subtilis carriers of the antibiotic resistant genes may have outcompeted the bacteria without the resistant plasmids (Figure 5).
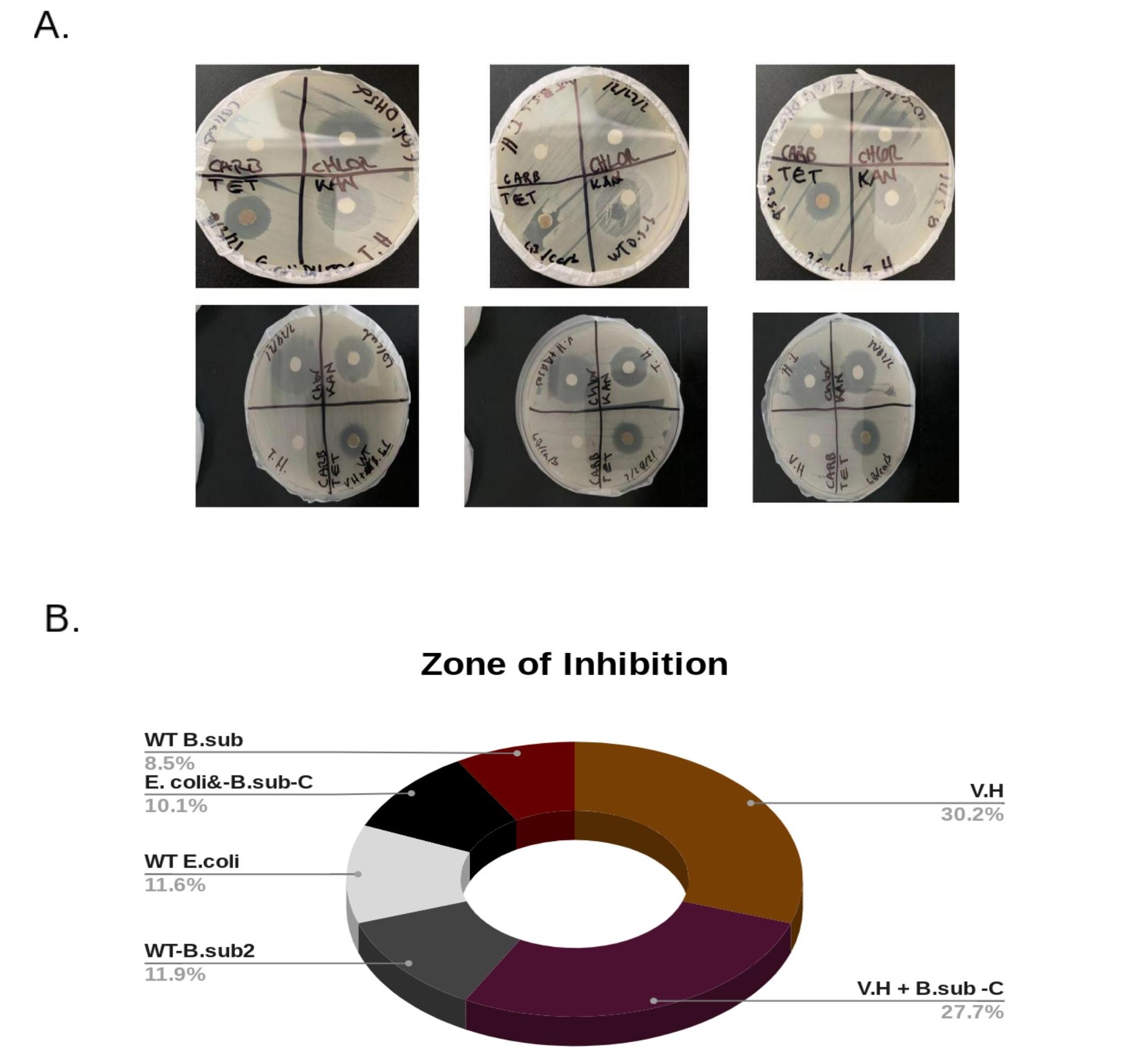
Figure 4. Antibiotic Resistance Tests.A. The top plates include the antibiotic resistant test results for E. Coli co-cultures and the bottom plates represent the results for V. harveyi sample groups. The top plates showed more moderate sensitivity to resistance of the antibiotics carbenicillin, chloramphenicol, tetracycline, and kanamycin. The WT E. Coli was more sensitive to tetracycline, chloramphenicol, and to kanamycin than the WT B. subtilis and the B.subtilis-Carrier-E. Coli co-culture. V. harveyi on the bottom plates showed identical and similar antibiotic resistance and sensitivity as the B.subtilis-Carrier-V.H and WT B. subtilis co-cultures. B. The zones of inhibition were measured in millimeters. The zone of inhibition for V. harveyi totaled 99 and 91 for B. subtilis-Carrier-V.H producing a 3.0% decrease in the zones of inhibition between each of these samples. The WT E. Coli had a total zone of inhibition of 8.5% that was raised to 10.1% for the B. subtilis-Carrier-E. Coli that yielded a total Zol of 33. The wild-type V. harveyi and E. Coli samples showed increased antibiotic sensitivity, however, the B. subtilis-Carrier-V.H and B. subtilis-Carrier-E. Coli co-cultures increased in antibiotic resistance.
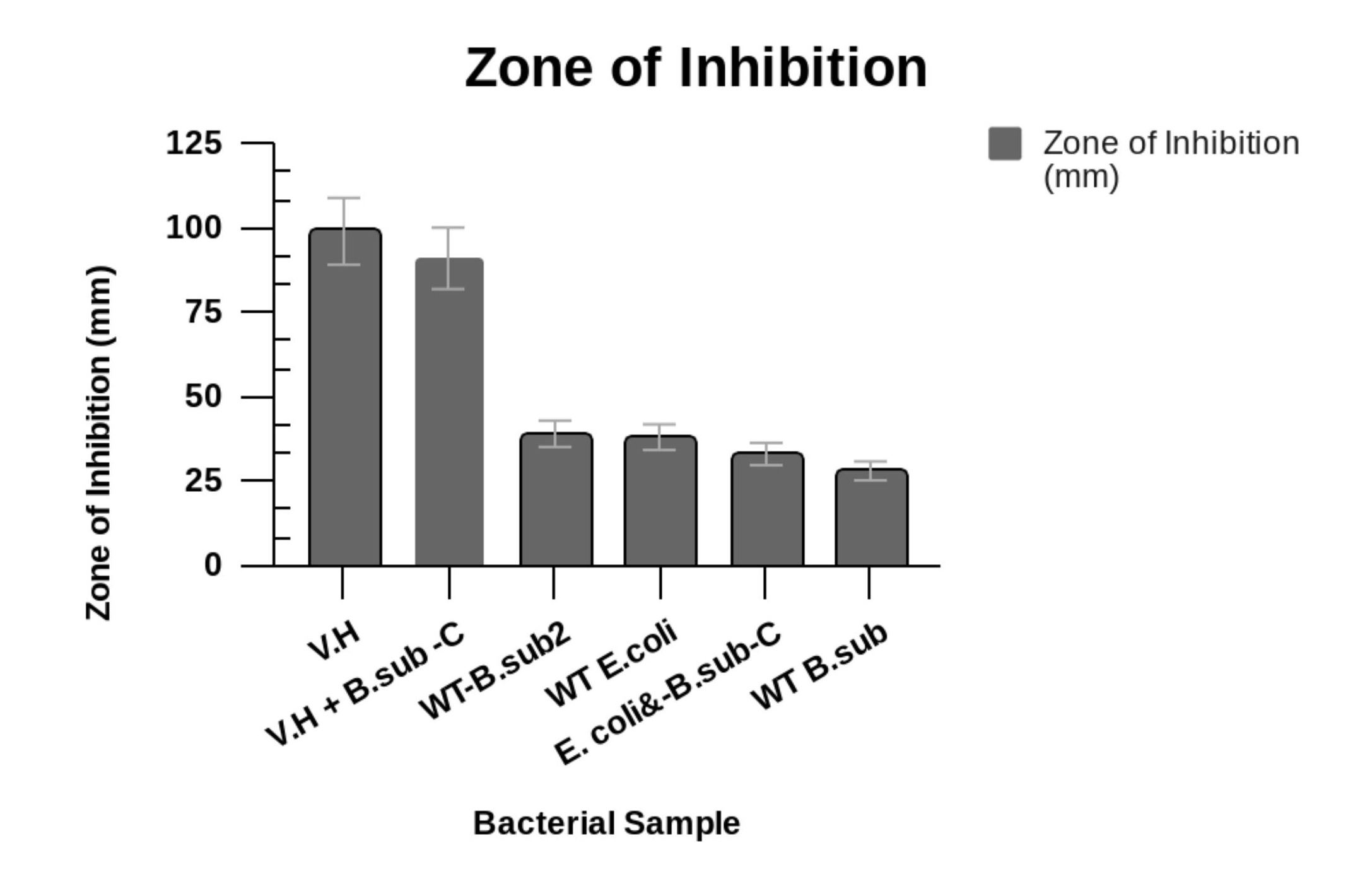
Figure 5. Zones of Inhibition (mm).The zones of inhibition were measured in millimeters. The zone of inhibition for V. harveyi totaled to 99 and 91 for B. subtilis-Carrier-V.H. WT E. Coli had a total zone of inhibition of 38 as B. subtilis-Carrier-E. Coli yielded a total Zol of 33. Wild-type V. harveyi and E. Coli samples showed increased antibiotic sensitivity, however, the B. subtilis Carrier-V.H and B. subtilis Carrier-E. Coli co-cultures increased in antibiotic resistance.
DISCUSSION
PLASMID CARRIAGE, BACTERIAL GROWTH, AND FITNESS
Many studies have shown that for plasmid persistence to occur there must be an association between plasmid loss, transfer through conjugation, and the effects of plasmid carriage on the fitness of the host. Gama et al., concluded that regardless of the multiple types of interactions affecting the persistence of plasmids, there is a more potent effect on plasmid maintenance than on those interactions impacting conjugation and plasmid loss [12, 13, 14, 15, 16, 17]. Additionally, Krone et al. used simulations of bacterial colony growth rates of plasmid carriers and plasmid-free strains as approximations for the growth rate parameters in their mathematical model [18]. They found a linear association between the radii of plasmid-carriers, plasmid-free bacteria, and cell growth rates [18]. As a result, in this study, the B. subtilis plasmid carrier co-cultures showed lesser levels of bacterial cell growth because plasmid carriage many times reduces the fitness of its host [5]. The frequency and rates of these dynamics depend on the plasmid, the environmental pressures, and on the host. Plasmid loss during bacterial growth can become sensed and detected [5]. For these reasons, the B.sub-C-VH and B.sub-C-E. coli co-cultures may have had lesser bacterial growth because of having a higher fitness cost. However, without plasmid loss or lacking any fitness cost, plasmids would still become removed from the populations of bacteria [5]. Plasmid persistence, thus, depends more on fitness cost and horizontal transfer via conjugation [5].
The fitness that is related to plasmid carriage immensely affects the plasmid persistence in bacterial populations [1, 2, 3, 4, 5]. However, the effects of plasmid fitness on wild-type bacterial host cells have not been completely surveyed [1, 2, 3, 4, 5]. Computational experiments have shown that fitness varies between bacterial strains in a community, which then increases the probability of plasmid persistence at low rates of conjugation [1, 2, 3, 4, 5]. For these reasons, the co-cultures with plasmid carriers may have shown higher fitness costs through displaying a bacterial growth reduction. In this study, the frequency of plasmid transfer may have been reduced as the amount of diverse bacterial strains in the cultures increased because the reception of plasmids was linked to a cost of fitness [1, 2, 3, 4, 5]. In addition, the acquisition of plasmids can be amplified when the allocation of fitness effects becomes more complex with a wider range of variance [1, 2, 3, 4, 5]. A possible interpretation of these effects is that the cost of plasmid fitness was similar between all bacterial strains in the cultures without plasmid carriers where higher rates of plasmid persistence occurred. The higher rates of plasmid persistence were observed in the cultures without carriers because there were lower rates of conjugation combined with lower fitness cost compared to the bacterial cultures with the more diverse bacterial strains, which required higher rates of conjugation [1, 2, 3, 4, 5]. For example, in the Benz et al. study they represented their model for the spread and proliferation of plasmids in a complex environment such as in the gut of animals and humans that predicted the rate of transfer using in vitro testing and genetic data analysis [19]. Benz et al. removed ESBL-plasmids from their native hosts, and they confirmed that the donor and recipient strains worked to regulate the frequencies of conjugation transfers [19]. However, the rate of conjugation needed for plasmid persistence may be much lower than confirmed in past studies [1, 2, 3, 4, 5]. An example includes a mathematical model that was created for the fitness effects of a carbapenem-resistance plasmid in 50 wild-type enterobacteria found in the gut microbiota of humans [1, 2, 3, 4, 5]. Alonso-Valle's mathematical model showed that the chance of plasmid persistence is not dependent on an escalated rate of conjugation. This effect yields plasmids at a low average cost, but forms incomplete plasmids at a larger cost of fitness [1, 2, 3, 4, 5]. The chance for plasmid persistence amplifies with the increased numbers of bacterial strains, creating a highly complex community [1, 2, 3, 4, 5]. As the community increases in complexity, the stability of plasmid proliferation also elevates through the transfer dynamics of the plasmids [1, 2, 3, 4, 5]. This in turn offers many available plasmid-permissive bacterial phylotypes [1, 2, 3, 4, 5].
In contrast, when the fitness effects differ, more available bacterial host cells in a population amplifies the chance of the uptake of plasmids by these host cells where no costs or favorable fitness effects are produced [1, 2, 3, 4, 5]. As a consequence, the B. sub-C-VH and the B.sub-C-E. coli co-cultures in this study may have decreased the availability of plasmid-free cells and exhibited reduced growth rates because of a greater exertion of a host fitness cost compared to the control samples. This evidence is significant because it confirms that plasmid carriage may have lessened the numbers of available plasmid-free bacterial cells needed for conserving host cell fitness and maintaining plasmid persistence. For instance, when plasmids are readily available in a complex bacterial community, plasmid persistence may continue even during the lack of conjugation [1, 2, 3, 4, 5]. Therefore, increasing the complexity of bacterial communities can increase the probability and continuance of plasmid persistence in natural environments [1, 2, 3, 4, 5]. Because many natural microbiomes consist of complex bacterial strains and communities, plasmids can easily proliferate within these natural environments [1]. Consequently, in this study, plasmid persistence may have occurred more frequently because of increased complexity present in the B. subtilis Carriers co-cultured with additional bacterial strains of V. harveyi and E. coli.
PLASMID CARRIAGE AND BIOFILM
More biofilm may have been produced in B.sub-C-VH treatment samples than in the B. sub-C-E. coli samples because of a greater presence of conjugative plasmids in the B.sub-C-VH samples than in the B.sub-C-E. coli cultures because conjugative plasmids improve the bacterial capability to form biofilms by using greater numbers of pili, which increases the adhesion within a biofilm [20]. Biofilm formation may have been higher in the B.sub-C-VH co-cultures than in the B.sub-C-E. coli because of the rate of conjugation that created more pilli formation. This increased pilli activity may have increased the adhesion in the biofilms of co-cultures with plasmid carriers. However, plasmids may still interplay with each other by organizing or inhibiting the transfer of plasmids [20]. Gama et al. examined and quantified the formation of biofilm in E. coli cells carrying two types of plasmids [20]. Gama et al. used 11 innate and extracted conjugative plasmids where they confirmed that a few plasmids promote the formation of biofilm through conjugation efficiency [20]. Gama et al. also confirmed the effect of paired plasmids on biofilm formation. Their study showed increased formation of biofilm when both plasmids inhabited the same bacterial cell or when the plasmids were contained and carried in different cells [20]. One of the plasmids showed dominance over the other plasmid where one of the plasmids directed the increase of biofilm formation. E. coli cells showed low formation of biofilm in a crystal violet biofilm assay versus in quinolone-resistance E. coli found in the same population [21]. Acquiring the two different plasmids of the E. coli QREC virulent strain lowered the production of biofilm in the microtiter plates [21]. The rates of motility were reduced, but planktonic growth continued to increase [21]. Therefore, Nesse et al. hypothesized that plasmids carrying antibiotic resistance genes may have reduced the formation of biofilm dependent upon the fluxes of flagellar protein expression [21]. Likewise, in this study, the co-cultures of E. coli combined with B. subtilis plasmid carriers, containing the resistance gene of ampicillin, showed less biofilm generation possibly due to a lesser production of flagellar protein expression from the E. coli cells [21]. Currently, methods are needed in future research studies to better interpret the molecular basis of these dynamic plasmid interactions [20].
Nesse et al. also observed that the acquisition of plasmids reduced the rate of motility in the transconjugants of the bacteria [21]. Earlier studies have confirmed this when the motility of E. coli strains increased after removing cephalosporin-resistance plasmids from the bacterial strains of E. coli [21]. The plasmids may have reduced the motility where the plasmids affected biofilm formation in the transconjugant bacterial cells by lowering the rate of motility [21]. These observations are supported by plasmids being acquired that did not affect the biofilm production on Congo Red (CR) agar plates, showing that the control of biofilm production depended on environmental conditions, which differed between the assays for biofilm [21]. The walls of the wells in a microtiter plate, at the liquid to air interface, of bacterial cells may have needed increased motility to reach the location of the biofilm [21]. This is opposite from the agar surface with biofilm formation because on agar surfaces intense motility levels may not be required for biofilm formation [21]. This is supported by previous studies of Salmonella flagellar mutants, in which these bacteria showed deformities in early biofilm creation in microtiter plates used for crystal violet biofilm tests [21]. In a subsequent study Salmonella displayed flagella that were not required for producing biofilm on CR-agar plates, but the lack of flagella did negatively affect the biofilm formed on air to liquid, surface to liquid, or cell to cell liquid interfaces [21]. Therefore, in this study, the E. coli cells co-cultured with B. subtilis carriers may have not shown profound biofilm formation because its plasmid carriage, containing ampicillin resistance genes, lowered the rate of motility needed for forming biofilm in the liquid cultures of the microtiter plates during earlier stages. This observation was depicted in the crystal violet biofilm assays of the B.sub-C-E Coli plasmid carriers, in which the B.sub-C-E. coli samples exhibited less biofilm formation possibly due to less motility negatively affecting the movement of the bacterial cells in the liquid to air interface during the early stage of biofilm generation.
However, in vibrios, biofilm creation is more robust because vibrios are highly motile during the early stages of biofilm formation in viscous liquid [22]. Although V. harveyi was co-cultured with the B. subtilis plasmid carriers, in this study, the B.sub-C-VH carrier cultures continued to show a greater intensity of biofilm formation because the V. harveyi produced a more vigorous motility. Many bacteria use their flagellum to produce much motility in the beginning stages of forming biofilm by increasing the movement into the biofilm or on its surface, but in vibrios motility involves more than initial attachment [22]. For example, it was shown in a previous study that Vibrio parahaemolyticus utilizes its motor to decide rather to move on a surface or swim in extremely viscous types of media [22]. V. parahaemolyticus uses its polar flagellum to initiate biofilm formation, so motility is central for biofilm formation in vibrios as observed in previous biofilm models [22]. The two flagellar systems power flagella through swimming in liquid media while the lateral flagellum drives the swarming on solid surfaces [23, 24, 25, 26, 27, 28]. The biosynthesis of the flagellar complexes is needed for swarm motility [29]. Bacterial cells from swarming colonies have more numbers of flagella than cells isolated from liquid cultures [29]. Wild-type B. subtilis are able to form biofilm complex structures on the surfaces of standing cultures of liquid [29]. However, researchers viewed deformities in these biofilm structures in B. subtilis mutants that lacked surfactin production [29]. B. subtilis produces surfactin as a self-defense mechanism. Bindel et al. found that because of a loss of swarming motility, their mutant B. subtilis sample of A164Δ10 could not form pellicles on their liquid cultures, but when they added proteases to the medium of A164Δ10 cultures, pellicles were produced [29]. The Bindel et al. study confirmed that there is a strong correlation between swarming motility and biofilm production [29]. Additionally, in this study, it may be inferred that the B. subtilis carriers and non-carriers co-cultured with E. coli and V. harveyi may have lacked biofilm formation because there may have been less surfactin production. In the tested samples of B.sub-C-E.coli and WT-B.sub-NC-VH, lesser amounts of surfactin may have led to a decreased rate of the B.subtilis ability to swarm, which then reduced the intensity of biofilm formed in the liquid culture crystal violet assays. Less biofilm was produced by the B.sub-C-E. coli growths due to the propensity of E. coli to have decreased rates of motility in liquid-to-air standing cultures while vibrios like V. harveyi use more vigorous flagellar motility in highly viscous medium not as utilized in liquid standing cultures. This reduction in swimming motility combined with a lesser presence and adhesion of E. coli cells or V. harveyi cells in the samples may have negatively affected the surfactin production of the B. subtilis plasmid carriers and of the wild-type B. subtilis non-carriers in these subsequent co-cultures, leading to the observed decrease of biofilm formation. Roder et al. showed that biofilms are spatial storages for plasmids, which allow plasmids to persist even in intense pressures of environmental conditions [30]. Bacterial cells that carry plasmids increase the removal of bacterial cells lacking plasmids where biofilms can become temporary storages for the plasmids [30]. However, plasmid maintenance expends much energy and leads to a fitness cost of the host cell [30].
PLASMID CARRIAGE AND ANTIBIOTIC RESISTANCE
Antibiotic resistance increased in the B.subtilis carrier co-cultures because plasmids can be transferred when their rate of conjugation and horizontal transfer overrides the costs of plasmid carriage in a bacterial cell [31]. In a previous study, Svara et al. constructed a mathematical model to study the dynamics of plasmids as they responded to varied treatments of antibiotics [31]. However, when there were reduced fitness costs of plasmids, a beneficial transfer of plasmids dwindled [31]. Svara et al. produced a model for the absence of non-resistant plasmids as plasmids remain in a bacterial population when the rate of transfer is most efficient and favourable [31]. Svara et al. found that the resistant plasmids, such as the plasmids with antibiotic resistance genes, dominated the non-resistant plasmids [31]. As a result, the plasmid carriers combined with the non-carriers, in this study, showed more antibiotic resistance. According to this previous study by Svara et al. these observations could have been a result of horizontal gene transfer where wild-type cells were more normally treated as the plasmids of wild-type bacteria that allow additional plasmids to enter bacterial cells [31]. Therefore, in a continuous presence of antibiotics, resistance genes in the host genome develop a high tolerance of treatments even in the presence of antibiotics.
Antibiotic resistance presents a pertinent problem for public health [31]. When resistance genes are transported through plasmid carriage or by phages, the spread can accelerate. Plasmids can allow the rapid spread of antibiotic resistance genes [31]. However, many pressures that favor the plasmid carriage of resistance genes have not been fully examined [31]. For plasmid maintenance in a bacterial population, bacteria carrying plasmids and the wild-type cells require the presence of plasmid-free bacterial cells [31]. A reason for this is that plasmid carriage combined with resistance has a cost of fitness that must be balanced with the availability of infection susceptible plasmid-free cells [31]. When the plasmid-free cells are eliminated by antibiotics, the plasmids have fewer cells to infect [31]. Hence, in this study, there may have been a higher fitness cost of the plasmid carriage in the B.sub-C-VH-E. coli samples because antibiotic resistance increased. The increased numbers of resistant bacterial cells outcompeted the reduced numbers of plasmid-free bacterial cells, including V. harveyi and E. coli, and this caused a high cost of fitness. However, Svara et al. showed that plasmids with lower transfer rates cannot carry antibiotic resistance genes [31].
As a consequence, our understanding of the conditions where antibiotic resistant genes are carried in plasmids can assist with developing strategies to lessen the proliferation and spread of resistance via blocking plasmid transfer [31]. Without the presence of antibiotic-mediated selection, bacteria can remove their resistant bacterial competitors when the resistance genes cause a high fitness cost [32, 33, 34, 35, 36, 37]. Conjugation and horizontal gene transfer allow for the ample persistence of plasmids, but an extremely high rate of conjugation is needed [38, 39, 40, 41, 42, 43, 44]. Conjugative plasmids can be transferred when there is a higher rate of conjugation remaining after the absence of antibiotics in E. coli samples [45, 46, 47, 48, 49, 50, 51]. The results from the Lopatkin et al. study showed that lessening the use of antibiotics is not sufficient enough for eliminating resistance [32, 33, 34, 35, 36, 37]. As a consequence, joining conjugation inhibition with increasing plasmid loss could become an improved method to lessen conjugation-dependent persistence of antibiotic resistance genes [32, 33, 34, 35, 36, 37].
HYPOTHETICAL MODEL FOR AN INTEGRATIVE CYCLE OF PLASMID PERSISTANCE
Plasmids can be shared and carry genes that assist bacterial hosts to survive in adverse and complex environments [1]. Additionally, when plasmids exhibit epistasis (fitness) and interact, the plasmids can be maintained in homeostasis in the population where this combination offers a benefit of fitness to the bacterial host cells [12, 13, 14, 15, 16, 17]. Plasmid-free cells can be non-present in the last and final bacterial population [12, 13, 14, 15, 16, 17]. Bacterial populations with one favourable plasmid did not have plasmid-free cells lacking in the final population [12, 13, 14, 15, 16, 17]. However, advantageous bacteria can still become extinct because only garnering fitness does not guarantee plasmid persistence [12]. Gama et al. results show that different assortments having positive epistasis can only survive and continue to proliferate when there is a lowered fitness cost, elevated rates of conjugation, and a decreased loss of transfer rates [12]. Contrarily, having low fitness cost, high rates of conjugation and reduced rates of loss is not a realistic condition for maintaining plasmid persistence [12, 13, 14, 15, 16, 17]. These three conditions are not probable because the rate of conjugation is dependent on the costs of fitness, as a result of increased expenditure of energy as well as metabolites where only plasmids can benefit from conjugation activity at a lower cost, but plasmids cannot persist in both of these two conditions, simultaneously. Thus, according to Roder et al., bacterial cells that carry plasmids increase the removal of bacterial cells lacking plasmids where biofilms can become temporary storages for plasmids [30]. Hence, Figure 6 shows a potential model for a cycle of plasmid persistence. The model is integrative in that it combines the three natural bacterial conditions, examined in this study, with host cell fitness and growth, conjugation, horizontal gene transfer (HGT), and the carriage of plasmids. The model may offer a potential visual explanation for the maintenance of plasmids in natural complex systems such as in the biofilm formed in an abiotic gut microbiome or in a biotic ecosystem.
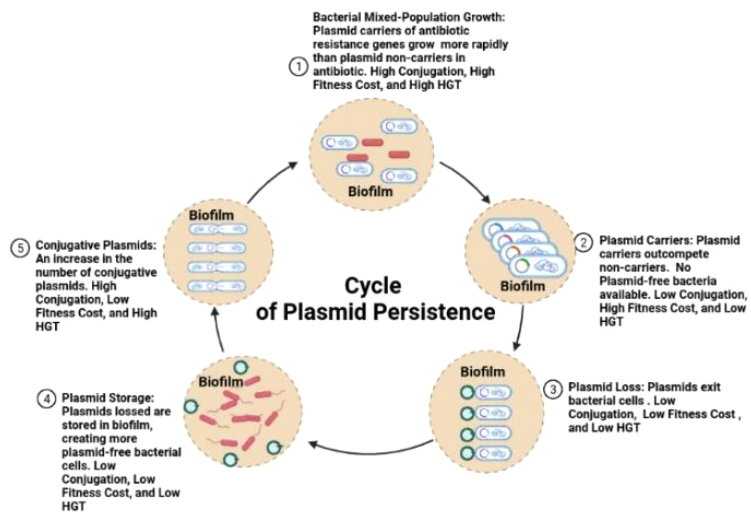
Figure 6. An integrative and Hypothetical Model for the Cycle of Plasmid Persistence.This hypothetical model shows a cycle for plasmid persistence in natural bacterial populations within communities of biofilm formed on abiotic or biotic surfaces. These bacterial communities begin growing with host cell plasmid carriers and non-carriers. The plasmid carriers grow more vigorously than the non-carriers until there are no plasmid-free bacteria available. At this point, there is a high fitness cost with low conjugation and low horizontal gene transfer (HGT). The plasmid carriers then begin losing plasmids at a high rate, and then the plasmids are stored in the biofilm. This generates more plasmid-free cells where the plasmids can then enter the cells. The level of conjugation heightens with a low fitness cost and a higher rate of HGT. Bacterial cells re-enter a state of growth to restart the cycle of plasmid persistence.
CONCLUSIONS
In this study, the natural phases of bacterial growth, biofilm production, and antibiotic resistance were examined in relation to plasmid carriage. These intrinsic stages combined with plasmid carriage were surveyed through in vitro co-culture assays in order to form a potential explanation for plasmid persistence in natural conditions. Major changes in these natural states were observed in the samples with the plasmid carriers versus the non-carriers. Bacterial growth decreased in all carrier co-cultures, biofilm was produced in B.subtilis carriers with V. harveyi, biofilm was reduced in B. subtilis carriers with E. coli, and the B.subtilis carrier V. harveyi and E. coli co-cultures increased in antibiotic resistance. It was concluded that plasmid carriage affects bacterial growth, biofilm formation, and the rate of antibiotic resistance, in which plasmid carriage, including each altered bacterial phase, contributes to plasmid persistence. However, a major limitation of this present study includes that the effects of plasmid fitness were only examined through in vitro methods of LB liquid culturing. This is a common practice in the field where past studies have displayed the effects of plasmid fitness with laboratory conditions that are associated with the same data measured in animal models [1, 2, 3, 4, 5]. As a result, the results in this present study may not completely represent the plasmid fitness in the human gut. Future studies are needed to further survey the total complexity of in vitro matrices with focused analysis of in vivo animal models. However, analyzing the cost of plasmid production in liquid cultures cannot represent the totality of bacterial dynamics in a spatial ecosystem, therefore, an image of observation created through a mathematical model of plasmid dynamics in bacterial biofilms and colonies is necessary for predicting the outcome of plasmids in bacterial populations [18]. Developing this model can be improved by including three dimensions such as multiple strains and multiple species of bacteria from many different ecosystems [18]. Including more dimensions can enhance our understanding and predictions for plasmids in spatial bacterial communities [18]. Through exploring these multi-dimensional microbial communities, more methods to stagnate the spread of plasmids can be found that may limit the propagation of antibiotic resistance.
ACKNOWLEDGEMENTS
Many thanks are given to my mentors who provided lectures, workshops, seminars, and research readings, which helped to establish my understanding of the fundamentals for microbiology. I also received hands-on experience in applying common and current novel microbiological techniques.














